The role of RNF149 in the pre-emptive quality control substrate ubiquitination
- PMID: 37031316
- PMCID: PMC10082771
- DOI: 10.1038/s42003-023-04763-9
The role of RNF149 in the pre-emptive quality control substrate ubiquitination
Abstract
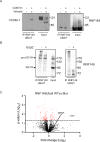
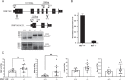
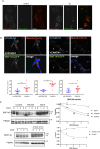
Protein quality control is a process in which a protein's folding status is constantly monitored. Mislocalized proteins (MLP), are processed by the various quality control pathways, as they are often misfolded due to inappropriate cellular surroundings. Polypeptides that fail to translocate into the ER due to an inefficient signal peptide, mutations or ER stress are recognized by the pre-emptive ER associated quality control (pEQC) pathway and degraded by the 26 S proteasome. In this report we reveal the role of RNF149, a membrane bound E3 ligase in the ubiquitination of known pEQC substrates. We demonstrate its selective binding only to non-translocated proteins and its association with known pEQC components. Impairment in RNF149 function increases translocation flux into the ER and manifests in a myeloproliferative neoplasm (MPN) phenotype, a pathological condition associated with pEQC impairment. Finally, the dynamic localization of RNF149 may provide a molecular switch to regulate pEQC during ER stress.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

