The role of native cysteine residues in the oligomerization of KCNQ1 channels
- PMID: 37031592
- PMCID: PMC10170711
- DOI: 10.1016/j.bbrc.2023.03.082
The role of native cysteine residues in the oligomerization of KCNQ1 channels
Abstract
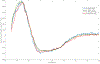
KCNQ1, the major component of the slow-delayed rectifier potassium channel, is responsible for repolarization of cardiac action potential. Mutations in this channel can lead to a variety of diseases, most notably long QT syndrome. It is currently unknown how many of these mutations change channel function and structure on a molecular level. Since tetramerization is key to proper function and structure of the channel, it is likely that mutations modify the stability of KCNQ1 oligomers. Presently, the C-terminal domain of KCNQ1 has been noted as the driving force for oligomer formation. However, truncated versions of this protein lacking the C-terminal domain still tetramerize. Therefore, we explored the role of native cysteine residues in a truncated construct of human KCNQ1, amino acids 100-370, by blocking potential interactions of cysteines with a nitroxide based spin label. Mobility of the spin labels was investigated with continuous wave electron paramagnetic resonance (CW-EPR) spectroscopy. The oligomerization state was examined by gel electrophoresis. The data provide information on tetramerization of human KCNQ1 without the C-terminal domain. Specifically, how blocking the side chains of native cysteines residues reduces oligomerization. A better understanding of tetramer formation could provide improved understanding of the molecular etiology of long QT syndrome and other diseases related to KCNQ1.
Keywords: EPR spectroscopy; KCNQ1; Kv channel; Oligomerization; SDS-PAGE.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

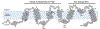
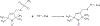
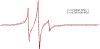
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

