Modulating auxin response stabilizes tomato fruit set
- PMID: 37032117
- PMCID: PMC10315294
- DOI: 10.1093/plphys/kiad205
Modulating auxin response stabilizes tomato fruit set
Erratum in
-
Correction: Alon Israeli and others, Modulating auxin response stabilizes tomato fruit set.Plant Physiol. 2023 Aug 3;192(4):3204. doi: 10.1093/plphys/kiad314. Plant Physiol. 2023. PMID: 37256789 Free PMC article. No abstract available.
Abstract
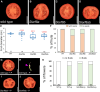
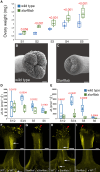
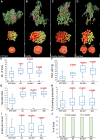
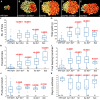
Fruit formation depends on successful fertilization and is highly sensitive to weather fluctuations that affect pollination. Auxin promotes fruit initiation and growth following fertilization. Class A auxin response factors (Class A ARFs) repress transcription in the absence of auxin and activate transcription in its presence. Here, we explore how multiple members of the ARF family regulate fruit set and fruit growth in tomato (Solanum lycopersicum) and Arabidopsis thaliana, and test whether reduction of SlARF activity improves yield stability in fluctuating temperatures. We found that several tomato Slarf mutant combinations produced seedless parthenocarpic fruits, most notably mutants deficient in SlARF8A and SlARF8B genes. Arabidopsis Atarf8 mutants deficient in the orthologous gene had less complete parthenocarpy than did tomato Slarf8a Slarf8b mutants. Conversely, Atarf6 Atarf8 double mutants had reduced fruit growth after fertilization. AtARF6 and AtARF8 likely switch from repression to activation of fruit growth in response to a fertilization-induced auxin increase in gynoecia. Tomato plants with reduced SlARF8A and SlARF8B gene dosage had substantially higher yield than the wild type under controlled or ambient hot and cold growth conditions. In field trials, partial reduction in the SlARF8 dose increased yield under extreme temperature with minimal pleiotropic effects. The stable yield of the mutant plants resulted from a combination of early onset of fruit set, more fruit-bearing branches and more flowers setting fruits. Thus, ARF8 proteins mediate the control of fruit set, and relieving this control with Slarf8 mutations may be utilized in breeding to increase yield stability in tomato and other crops.
© American Society of Plant Biologists 2023. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. NO and AI are inventors in a Provisional patent application No. 63/267,407, INCREASING YIELD STABILITY IN PLANTS that includes data described in this paper.
Figures



References
-
- Abad M, Monteiro AA. The use of auxins for the production of greenhouse tomatoes in mild-winter conditions: a review. Sci Hortic. 1989:38(3–4):167–192. 10.1016/0304-4238(89)90064-2 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

