Super-enhancer-driven TOX2 mediates oncogenesis in Natural Killer/T Cell Lymphoma
- PMID: 37032358
- PMCID: PMC10084643
- DOI: 10.1186/s12943-023-01767-1
Super-enhancer-driven TOX2 mediates oncogenesis in Natural Killer/T Cell Lymphoma
Abstract
Background: Extranodal natural killer/T-cell lymphoma (NKTL) is an aggressive type of non-Hodgkin lymphoma with dismal outcome. A better understanding of disease biology and key oncogenic process is necessary for the development of targeted therapy. Super-enhancers (SEs) have been shown to drive pivotal oncogenes in various malignancies. However, the landscape of SEs and SE-associated oncogenes remain elusive in NKTL.
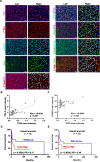
Methods: We used Nano-ChIP-seq of the active enhancer marker histone H3 lysine 27 acetylation (H3K27ac) to profile unique SEs NKTL primary tumor samples. Integrative analysis of RNA-seq and survival data further pinned down high value, novel SE oncogenes. We utilized shRNA knockdown, CRISPR-dCas9, luciferase reporter assay, ChIP-PCR to investigate the regulation of transcription factor (TF) on SE oncogenes. Multi-color immunofluorescence (mIF) staining was performed on an independent cohort of clinical samples. Various function experiments were performed to evaluate the effects of TOX2 on the malignancy of NKTL in vitro and in vivo.
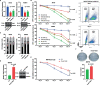
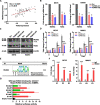
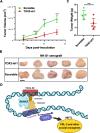
Results: SE landscape was substantially different in NKTL samples in comparison with normal tonsils. Several SEs at key transcriptional factor (TF) genes, including TOX2, TBX21(T-bet), EOMES, RUNX2, and ID2, were identified. We confirmed that TOX2 was aberrantly overexpressed in NKTL relative to normal NK cells and high expression of TOX2 was associated with worse survival. Modulation of TOX2 expression by shRNA, CRISPR-dCas9 interference of SE function impacted on cell proliferation, survival and colony formation ability of NKTL cells. Mechanistically, we found that RUNX3 regulates TOX2 transcription by binding to the active elements of its SE. Silencing TOX2 also impaired tumor formation of NKTL cells in vivo. Metastasis-associated phosphatase PRL-3 has been identified and validated as a key downstream effector of TOX2-mediated oncogenesis.
Conclusions: Our integrative SE profiling strategy revealed the landscape of SEs, novel targets and insights into molecular pathogenesis of NKTL. The RUNX3-TOX2-SE-TOX2-PRL-3 regulatory pathway may represent a hallmark of NKTL biology. Targeting TOX2 could be a valuable therapeutic intervene for NKTL patients and warrants further study in clinic.
Keywords: Epigenetics; Natural Killer/T Cell Lymphoma; PRL-3; RUNX3; Super-enhancer; TOX2; Therapeutic targets.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

