Species identification, antibiotic resistance, and virulence in Enterobacter cloacae complex clinical isolates from South Korea
- PMID: 37032871
- PMCID: PMC10076837
- DOI: 10.3389/fmicb.2023.1122691
Species identification, antibiotic resistance, and virulence in Enterobacter cloacae complex clinical isolates from South Korea
Abstract
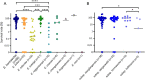
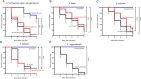
This study aimed to identify the species of Enterobacter cloacae complex (ECC) isolates and compare the genotype, antibiotic resistance, and virulence among them. A total of 183 ECC isolates were collected from patients in eight hospitals in South Korea. Based on partial sequences of hsp60 and phylogenetic analysis, all ECC isolates were identified as nine species and six subspecies. Enterobacter hormaechei was the predominant species (47.0%), followed by Enterobacter kobei, Enterobacter asburiae, Enterobacter ludiwigii, and Enterobacter roggenkampii. Multilocus sequence typing analysis revealed that dissemination was not limited to a few clones, but E. hormaechei subsp. xiangfangensis, E. hormaechei subsp. steigerwaltii, and E. ludwigii formed large clonal complexes. Antibiotic resistance rates were different between the ECC species. In particular, E. asburiae, E. kobei, E. roggenkampii, and E. cloacae isolates were highly resistant to colistin, whereas most E. hormaechei and E. ludwigii isolates were susceptible to colistin. Virulence was evaluated through serum bactericidal assay and the Galleria mellonella larvae infection model. Consistency in the results between the serum resistance and the G. mellonella larvae infection assay was observed. Serum bactericidal assay showed that E. hormaechei, E. kobei, and E. ludwigii were significantly more virulent than E. asburiae and E. roggenkampii. In this study, we identified the predominant ECC species in South Korea and observed the differences in antibiotic resistance and virulence between the species. Our findings suggest that correct species identification, as well as continuous monitoring is crucial in clinical settings.
Keywords: Enterobacter cloacae complex; Enterobacter hormaechei; antibiotic resistance; species identification; virulence.
Copyright © 2023 Ganbold, Seo, Wi, Kwon and Ko.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Clinical and laboratory standards institute (CLSI) . Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Ninth Informational Supplement M100-S31. (Wayne, PA: CLSI; ). (2021)
-
- Di Pilato V., Henrici De Angelis L., Aiezza N., Baccani I., Niccolai C., Parisio E. M., et al. . (2022). Resistome and virulome accretion in an NDM-1- producing ST147 sublineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: a genotypic and phenotypic characterisation. Lancet Microbe. 3, e224–e234. doi: 10.1016/S2666-5247(21)00268-8, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

