First report of coexistence of bla KPC-2-, bla NDM-1- and mcr-9-carrying plasmids in a clinical carbapenem-resistant Enterobacter hormaechei isolate
- PMID: 37032905
- PMCID: PMC10076803
- DOI: 10.3389/fmicb.2023.1153366
First report of coexistence of bla KPC-2-, bla NDM-1- and mcr-9-carrying plasmids in a clinical carbapenem-resistant Enterobacter hormaechei isolate
Abstract
Introduction: Colistin is regarded as one of the last-resort antibiotics against severe infections caused by carbapenem-resistant Enterobacteriaceae. Strains with cooccurrence of mcr-9 and carbapenemase genes are of particular concern. This study aimed to investigate the genetic characteristics of a bla KPC-2-carrying plasmid, bla NDM-1-carrying plasmid and mcr-9-carrying plasmid coexisting in a carbapenem-resistant Enterobacter hormaechei isolate.
Methods: E. hormaechei strain E1532 was subjected to whole-genome sequencing, and the complete nucleotide sequences of three resistance plasmids identified in the strain were compared with related plasmid sequences. The resistance phenotypes mediated by these plasmids were analyzed by plasmid transfer, carbapenemase activity and antimicrobial susceptibility testing.
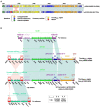
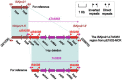
Results: Whole-genome sequencing revealed that strain E1532 carries three different resistance plasmids, pE1532-KPC, pE1532-NDM and pE1532-MCR. pE1532-KPC harboring bla KPC-2 and pE1532-NDM harboring bla NDM-1 are highly identical to the IncR plasmid pHN84KPC and IncX3 plasmid pNDM-HN380, respectively. The mcr-9-carrying plasmid pE1532-MCR possesses a backbone highly similar to that of the IncHI2 plasmids R478 and p505108-MDR, though their accessory modules differ. These three coexisting plasmids carry a large number of resistance genes and contribute to high resistance to almost all antibiotics tested, except for amikacin, trimethoprim/sulfamethoxazole, tigecycline and polymyxin B. Most of the plasmid-mediated resistance genes are located in or flanked by various mobile genetic elements, facilitating horizontal transfer of antibiotic resistance genes.
Discussion: This is the first report of a single E. hormaechei isolate with coexistence of three resistance plasmids carrying mcr-9 and the two most common carbapenemase genes, bla KPC-2 and bla NDM-1. The prevalence and genetic features of these coexisting plasmids should be monitored to facilitate the establishment of effective strategies to control their further spread.
Keywords: Enterobacter hormaechei; carbapenemase genes; mcr-9; multidrug resistance; plasmid.
Copyright © 2023 Yuan, Xia, Xiong, Xie, Lv, Sun and Feng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Borowiak M., Baumann B., Fischer J., Thomas K., Deneke C., Hammerl J. A., et al. (2020). Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in Colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011-2018) in Germany. Front. Microbiol. 11:80. doi: 10.3389/fmicb.2020.00080, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

