Transcriptome analysis of Lr19-virulent mutants provides clues for the AvrLr19 of Puccinia triticina
- PMID: 37032911
- PMCID: PMC10073493
- DOI: 10.3389/fmicb.2023.1062548
Transcriptome analysis of Lr19-virulent mutants provides clues for the AvrLr19 of Puccinia triticina
Abstract
Introduction: Wheat leaf rust caused by Puccinia triticina (Pt) remains one of the most destructive diseases of common wheat worldwide. Understanding the pathogenicity mechanisms of Pt is important to control wheat leaf rust.
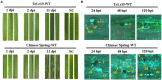
Methods: The urediniospores of Pt race PHNT (wheat leaf rust resistance gene Lr19-avirulent isolate) were mutagenized with ethyl methanesulfonate (EMS), and two Lr19-virulent mutants named M1 and M2 were isolated. RNA sequencing was performed on samples collected from wheat cultivars Chinese Spring and TcLr19 infected with wild-type (WT) PHNT, M1, and M2 isolates at 14 days post-inoculation (dpi), respectively. Screening AvrLr19 candidates by quantitative reverse transcription PCR (qPCR) and Agrobacterium-mediated transient assays in Nicotiana benthamiana.
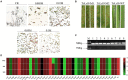
Results: 560 genes with single nucleotide polymorphisms (SNPs) and insertions or deletions (Indels) from non-differentially expressed genes were identified. Among them, 10 secreted proteins were screened based on their fragments per kilobase of exon model per million mapped reads (FPKM) values in the database. qPCR results showed that the expression profiles of 7 secreted proteins including PTTG_27471, PTTG_12441, PTTG_28324, PTTG_26499, PTTG_06910, PTTG_26516, and PTTG_03570 among 10 secreted proteins in mutants were significantly different with that in wild-type isolate after infection wheat TcLr19 and might be related to the recognition between Lr19 and AvrLr19. In addition, a total of 216 differentially expressed genes (DEGs) were obtained from three different sample comparisons including M1-vs-WT, M2-vs-WT, and M1-vs-M2. Among 216 DEGs, 15 were predicted to be secreted proteins. One secreted protein named PTTG_04779 could inhibit programmed progress of cell death (PCD) induced by apoptosis-controlling genes B-cell lymphoma-2 associated X protein (BAX) on Nicotiana benthamiana, indicating that it might play a virulence function in plant. Taken together, total 8 secreted proteins, PTTG_04779, PTTG_27471, PTTG_12441, PTTG_28324, PTTG_26499, PTTG_06910, PTTG_26516, PTTG_03570 are identified as AvrLr19 candidates.
Discussion: Our results showed that a large number of genes participate in the interaction between Pt and TcLr19, which will provide valuable resources for the identification of AvrLr19 candidates and pathogenesis-related genes.
Keywords: AvrLr19 candidates; Puccinia triticina; sequence insertion or deletion; single nucleotide polymorphism; wheat.
Copyright © 2023 Cui, Wu, Fan, Wang, Liu, Di and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Genome-Wide Identification of Effector Candidates With Conserved Motifs From the Wheat Leaf Rust Fungus Puccinia triticina.Front Microbiol. 2020 Jun 3;11:1188. doi: 10.3389/fmicb.2020.01188. eCollection 2020. Front Microbiol. 2020. PMID: 32582112 Free PMC article.
-
A wheat NBS-LRR gene TaRGA19 participates in Lr19-mediated resistance to Puccinia triticina.Plant Physiol Biochem. 2017 Oct;119:1-8. doi: 10.1016/j.plaphy.2017.08.009. Epub 2017 Aug 15. Plant Physiol Biochem. 2017. PMID: 28837844
-
Lr19 Resistance in Wheat Becomes Susceptible to Puccinia triticina in India.Plant Dis. 2005 Dec;89(12):1360. doi: 10.1094/PD-89-1360A. Plant Dis. 2005. PMID: 30791320
-
The progress of leaf rust research in wheat.Fungal Biol. 2020 Jun;124(6):537-550. doi: 10.1016/j.funbio.2020.02.013. Epub 2020 Feb 29. Fungal Biol. 2020. PMID: 32448445 Review.
-
Wheat leaf rust caused by Puccinia triticina.Mol Plant Pathol. 2008 Sep;9(5):563-75. doi: 10.1111/j.1364-3703.2008.00487.x. Mol Plant Pathol. 2008. PMID: 19018988 Free PMC article. Review.
Cited by
-
Wheat Leaf Rust Effector Pt48115 Localized in the Chloroplasts and Suppressed Wheat Immunity.J Fungi (Basel). 2025 Jan 20;11(1):80. doi: 10.3390/jof11010080. J Fungi (Basel). 2025. PMID: 39852499 Free PMC article.
-
The Underexplored Mechanisms of Wheat Resistance to Leaf Rust.Plants (Basel). 2023 Nov 28;12(23):3996. doi: 10.3390/plants12233996. Plants (Basel). 2023. PMID: 38068630 Free PMC article. Review.
-
Wheat Leaf Rust Fungus Effector Protein Pt1641 Is Avirulent to TcLr1.Plants (Basel). 2024 Aug 14;13(16):2255. doi: 10.3390/plants13162255. Plants (Basel). 2024. PMID: 39204691 Free PMC article.
-
Evasion of wheat resistance gene Lr15 recognition by the leaf rust fungus is attributed to the coincidence of natural mutations and deletion in AvrLr15 gene.Mol Plant Pathol. 2024 Jul;25(7):e13490. doi: 10.1111/mpp.13490. Mol Plant Pathol. 2024. PMID: 38952297 Free PMC article.
References
-
- Anderson C., Khan M. A., Catanzariti A. M., Jack C. A., Nemri A., Lawrence G. J., et al. . (2016). Genome analysis and avirulence gene cloning using a high-density RADseq linkage map of the flax rust fungus, Melampsora lini. BMC Genom. 17, 667–687. doi: 10.1186/s12864-016-3011-9, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

