Computer-aided genomic data analysis of drug-resistant Neisseria gonorrhoeae for the Identification of alternative therapeutic targets
- PMID: 37033487
- PMCID: PMC10080061
- DOI: 10.3389/fcimb.2023.1017315
Computer-aided genomic data analysis of drug-resistant Neisseria gonorrhoeae for the Identification of alternative therapeutic targets
Abstract
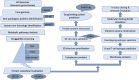
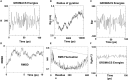
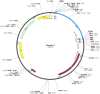
Neisseria gonorrhoeae is an emerging multidrug resistance pathogen that causes sexually transmitted infections in men and women. The N. gonorrhoeae has demonstrated an emerging antimicrobial resistance against reported antibiotics, hence fetching the attention of researchers to address this problem. The present in-silico study aimed to find putative novel drug and vaccine targets against N. gonorrhoeae infection by the application of bioinformatics approaches. Core genes set of 69 N. gonorrhoeae strains was acquired from complete genome sequences. The essential and non-homologous metabolic pathway proteins of N. gonorrhoeae were identified. Moreover, different bioinformatics databases were used for the downstream analysis. The DrugBank database scanning identified 12 novel drug targets in the prioritized list. They were preferred as drug targets against this bacterium. A viable vaccine is unavailable so far against N. gonorrhoeae infection. In the current study, two outer-membrane proteins were prioritized as vaccine candidates via reverse vaccinology approach. The top lead B and T-cells overlapped epitopes were utilized to generate a chimeric vaccine construct combined with immune-modulating adjuvants, linkers, and PADRE sequences. The top ranked prioritized vaccine construct (V7) showed stable molecular interaction with human immune cell receptors as inferred during the molecular docking and MD simulation analyses. Considerable response for immune cells was interpreted by in-silico immune studies. Additional tentative validation is required to ensure the effectiveness of the prioritized vaccine construct against N. gonorrhoeae infection. The identified proteins can be used for further rational drug and vaccine designing to develop potential therapeutic entities against the multi-drug resistant N. gonorrhoeae.
Keywords: Neisseria gonorrhoeae; chimaric vaccine; epitope; multi-drug resistant; novel drug target.
Copyright © 2023 Qasim, Jaan, Wara, Shehroz, Nishan, Shams, Shah and Ojha.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Reverse vaccinology approaches to introduce promising immunogenic and drug targets against antibiotic-resistant Neisseria gonorrhoeae: Thinking outside the box in current prevention and treatment.Infect Genet Evol. 2023 Aug;112:105449. doi: 10.1016/j.meegid.2023.105449. Epub 2023 May 22. Infect Genet Evol. 2023. PMID: 37225067
-
Characterization of exclusively non-commensal Neisseria gonorrhoeae pangenome to prioritize globally conserved and thermodynamically stable vaccine candidates using immune-molecular dynamic simulations.Microb Pathog. 2023 Dec;185:106439. doi: 10.1016/j.micpath.2023.106439. Epub 2023 Nov 7. Microb Pathog. 2023. PMID: 37944674
-
Genetic Similarity of Gonococcal Homologs to Meningococcal Outer Membrane Proteins of Serogroup B Vaccine.mBio. 2019 Sep 10;10(5):e01668-19. doi: 10.1128/mBio.01668-19. mBio. 2019. PMID: 31506309 Free PMC article.
-
Applications of genomics to slow the spread of multidrug-resistant Neisseria gonorrhoeae.Ann N Y Acad Sci. 2019 Jan;1435(1):93-109. doi: 10.1111/nyas.13871. Epub 2018 Jun 6. Ann N Y Acad Sci. 2019. PMID: 29876934 Free PMC article. Review.
-
Neisseria gonorrhoeae vaccines: a contemporary overview.Clin Microbiol Rev. 2024 Mar 14;37(1):e0009423. doi: 10.1128/cmr.00094-23. Epub 2024 Jan 16. Clin Microbiol Rev. 2024. PMID: 38226640 Free PMC article. Review.
Cited by
-
Structural informatics approach for designing an epitope-based vaccine against the brain-eating Naegleria fowleri.Front Immunol. 2023 Oct 30;14:1284621. doi: 10.3389/fimmu.2023.1284621. eCollection 2023. Front Immunol. 2023. PMID: 37965306 Free PMC article.
-
Genome-level therapeutic targets identification and chimeric Vaccine designing against the Blastomyces dermatitidis.Heliyon. 2024 Aug 14;10(16):e36153. doi: 10.1016/j.heliyon.2024.e36153. eCollection 2024 Aug 30. Heliyon. 2024. PMID: 39224264 Free PMC article.
-
Deciphering the Immunogenicity of Monkeypox Proteins for Designing the Potential mRNA Vaccine.ACS Omega. 2023 Oct 30;8(45):43341-43355. doi: 10.1021/acsomega.3c07866. eCollection 2023 Nov 14. ACS Omega. 2023. PMID: 38024731 Free PMC article.
-
Addressing Sexually Transmitted Infections Due to Neisseria gonorrhoeae in the Present and Future.Microorganisms. 2024 Apr 28;12(5):884. doi: 10.3390/microorganisms12050884. Microorganisms. 2024. PMID: 38792714 Free PMC article. Review.
-
Proteome level analysis of drug-resistant Prevotella melaninogenica for the identification of novel therapeutic candidates.Front Microbiol. 2023 Sep 22;14:1271798. doi: 10.3389/fmicb.2023.1271798. eCollection 2023. Front Microbiol. 2023. PMID: 37808310 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

