Nucleophile responsive charge-reversing polycations for pDNA transfection
- PMID: 37033743
- PMCID: PMC10071491
- DOI: 10.1039/d3py00075c
Nucleophile responsive charge-reversing polycations for pDNA transfection
Abstract
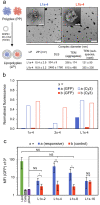
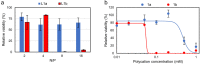
Polycationic carriers promise low cost and scalable gene therapy treatments, however inefficient intracellular unpacking of the genetic cargo has limited transfection efficiency. Charge-reversing polycations, which transition from cationic to neutral or negative charge, can offer targeted intracellular DNA release. We describe a new class of charge-reversing polycation which undergoes a cationic-to-neutral conversion by a reaction with cellular nucleophiles. The deionization reaction is relatively slow with primary amines, and much faster with thiols. In mammalian cells, the intracellular environment has elevated concentrations of amino acids (∼10×) and the thiol glutathione (∼1000×). We propose this allows for decationization of the polymeric carrier slowly in the extracellular space and then rapidly in the intracellular milleu for DNA release. We demonstrate that in a lipopolyplex formulation this leads to both improved transfection and reduced cytotoxicity when compared to a non-responsive polycationic control.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts of interest.
Figures




Similar articles
-
pH-responsive Multi-PEGylated dual cationic nanoparticles enable charge modulations for safe gene delivery.ChemMedChem. 2007 Sep;2(9):1321-7. doi: 10.1002/cmdc.200700093. ChemMedChem. 2007. PMID: 17579918
-
Supramolecular host-guest polycationic gene delivery system based on poly(cyclodextrin) and azobenzene-terminated polycations.Colloids Surf B Biointerfaces. 2016 Nov 1;147:25-35. doi: 10.1016/j.colsurfb.2016.07.028. Epub 2016 Jul 25. Colloids Surf B Biointerfaces. 2016. PMID: 27478960
-
Block catiomer polyplexes with regulated densities of charge and disulfide cross-linking directed to enhance gene expression.J Am Chem Soc. 2004 Mar 3;126(8):2355-61. doi: 10.1021/ja0379666. J Am Chem Soc. 2004. PMID: 14982439
-
Degradable branched polycationic systems for nucleic acid delivery.Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2020 Sep;12(5):e1631. doi: 10.1002/wnan.1631. Epub 2020 Mar 16. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2020. PMID: 32180362 Review.
-
Polycation-Based Ternary Gene Delivery System.Curr Drug Metab. 2015;16(2):152-65. doi: 10.2174/138920021602150713115608. Curr Drug Metab. 2015. PMID: 26179609 Review.
Cited by
-
Chemical reaction networks based on conjugate additions on β'-substituted Michael acceptors.Chem Commun (Camb). 2023 Sep 19;59(75):11174-11187. doi: 10.1039/d3cc02126b. Chem Commun (Camb). 2023. PMID: 37529876 Free PMC article. Review.
-
Poly(l-glutamic acid) augments the transfection performance of lipophilic polycations by overcoming tradeoffs among cytotoxicity, pDNA delivery efficiency, and serum stability.RSC Appl Polym. 2024 Apr 30;2(4):701-718. doi: 10.1039/d4lp00085d. eCollection 2024 Jul 18. RSC Appl Polym. 2024. PMID: 39035825 Free PMC article.
References
LinkOut - more resources
Full Text Sources
