Development of three-dimensional primary human myospheres as culture model of skeletal muscle cells for metabolic studies
- PMID: 37034250
- PMCID: PMC10076718
- DOI: 10.3389/fbioe.2023.1130693
Development of three-dimensional primary human myospheres as culture model of skeletal muscle cells for metabolic studies
Erratum in
-
Correction: Development of three-dimensional primary human myospheres as culture model of skeletal muscle cells for metabolic studies.Front Bioeng Biotechnol. 2025 Sep 16;13:1687822. doi: 10.3389/fbioe.2025.1687822. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 41036384 Free PMC article.
Abstract
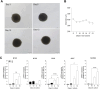
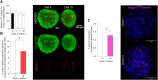
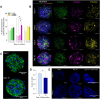
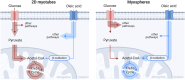
Introduction: Skeletal muscle is a major contributor to whole-body energy homeostasis and the utilization of fatty acids and glucose. At present, 2D cell models have been the most used cellular models to study skeletal muscle energy metabolism. However, the transferability of the results to in vivo might be limited. This project aimed to develop and characterize a skeletal muscle 3D cell model (myospheres) as an easy and low-cost tool to study molecular mechanisms of energy metabolism. Methods and results: We demonstrated that human primary myoblasts form myospheres without external matrix support and carry structural and molecular characteristics of mature skeletal muscle after 10 days of differentiation. We found significant metabolic differences between the 2D myotubes model and myospheres. In particular, myospheres showed increased lipid oxidative metabolism than the 2D myotubes model, which oxidized relatively more glucose and accumulated more oleic acid. Discussion and conclusion: These analyses demonstrate model differences that can have an impact and should be taken into consideration for studying energy metabolism and metabolic disorders in skeletal muscle.
Keywords: 3D cell model; energy metabolism; metabolic disorders; muscle spheroid; myosphere; skeletal muscle.
Copyright © 2023 Dalmao-Fernandez, Aizenshtadt, Bakke, Krauss, Rustan, Thoresen and Kase.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Al-Khalili L., Chibalin A. V., Kannisto K., Zhang B. B., Permert J., Holman G. D., et al. (2003). Insulin action in cultured human skeletal muscle cells during differentiation: Assessment of cell surface GLUT4 and GLUT1 content. Cell Mol. Life Sci. 60, 991–998. 10.1007/s00018-003-3001-3 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

