High Expression of Heterogeneous Nuclear Ribonucleoprotein A1 Facilitates Hepatocellular Carcinoma Growth
- PMID: 37034304
- PMCID: PMC10075271
- DOI: 10.2147/JHC.S402247
High Expression of Heterogeneous Nuclear Ribonucleoprotein A1 Facilitates Hepatocellular Carcinoma Growth
Abstract
Purpose: Hepatocellular carcinoma (HCC) represents one of the most common tumors in the world. Our study aims to explore new markers and therapeutic targets for HCC. Heterogeneous Nuclear ribonucleoprotein A1 (hnRNPA1) has recently been found to be involved in the progression of several types of cancer, but its role in HCC remains uncovered.
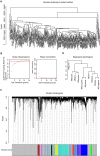
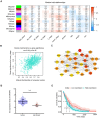
Methods: We performed bioinformatic analysis to preliminarily show the relationship between hnRNPA1 and liver cancer. Then the correlation of the hnRNPA1 gene expression with clinicopathological characteristics of HCC patients was verified by human liver cancer tissue microarrays. The functional role of this gene was evaluated by in vivo and vitro experiments.
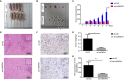
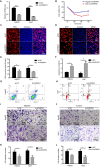
Results: Results showed that the expression of hnRNPA1 was upregulated in HCC tissues and was associated with pathological stage of HCC patients. Knockdown of hnRNPA1 gene markedly inhibited tumor growth in vivo, and reversed the effects on proliferation, migration and invasion and promoted apoptosis in vitro. Furthermore, down-regulation of hnRNPA1 gene expression can inhibit the activity of the MEK/ERK pathway.
Conclusion: In our work, we combined bioinformatic analysis with in vivo and in vitro experiments to initially elucidate the function of hnRNPA1 in liver cancer, which may help to explore biomarkers and therapeutic targets for HCC patients.
Keywords: MEK/ERK; WGCNA; hepatocellular carcinoma; hnRNPA1; proliferation.
© 2023 Cao et al.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
LncRNA ANCR promotes hepatocellular carcinoma metastasis through upregulating HNRNPA1 expression.RNA Biol. 2020 Mar;17(3):381-394. doi: 10.1080/15476286.2019.1708547. Epub 2020 Jan 2. RNA Biol. 2020. PMID: 31868085 Free PMC article.
-
Prognostic value and oncogene function of heterogeneous nuclear ribonucleoprotein A1 overexpression in HBV-related hepatocellular carcinoma.Int J Biol Macromol. 2019 May 15;129:140-151. doi: 10.1016/j.ijbiomac.2019.02.012. Epub 2019 Feb 4. Int J Biol Macromol. 2019. PMID: 30731163
-
Prognostic value of heterogeneous ribonucleoprotein A1 expression and inflammatory indicators for patients with surgically resected hepatocellular carcinoma: Perspectives from a high occurrence area of hepatocellular carcinoma in China.Oncol Lett. 2018 Sep;16(3):3746-3756. doi: 10.3892/ol.2018.9079. Epub 2018 Jul 4. Oncol Lett. 2018. PMID: 30127985 Free PMC article.
-
Downregulation of hnRNPA1 inhibits hepatocellular carcinoma cell progression by modulating alternative splicing of ZNF207 exon 9.Front Oncol. 2025 Jan 6;14:1517459. doi: 10.3389/fonc.2024.1517459. eCollection 2024. Front Oncol. 2025. PMID: 39834948 Free PMC article.
-
High expression of hnRNPA1 promotes cell invasion by inducing EMT in gastric cancer.Oncol Rep. 2018 Apr;39(4):1693-1701. doi: 10.3892/or.2018.6273. Epub 2018 Feb 16. Oncol Rep. 2018. PMID: 29484423 Free PMC article.
Cited by
-
hnRNPA1 promotes the metastasis and proliferation of gastric cancer cells through WISP2-guided Wnt/β-catenin signaling pathway.Discov Oncol. 2024 Sep 19;15(1):465. doi: 10.1007/s12672-024-01354-w. Discov Oncol. 2024. PMID: 39298013 Free PMC article.
-
The Roles of hnRNP Family in the Brain and Brain-Related Disorders.Mol Neurobiol. 2024 Jun;61(6):3578-3595. doi: 10.1007/s12035-023-03747-4. Epub 2023 Nov 24. Mol Neurobiol. 2024. PMID: 37999871 Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

