The phytase RipBL1 enables the assignment of a specific inositol phosphate isomer as a structural component of human kidney stones
- PMID: 37034402
- PMCID: PMC10074554
- DOI: 10.1039/d2cb00235c
The phytase RipBL1 enables the assignment of a specific inositol phosphate isomer as a structural component of human kidney stones
Abstract
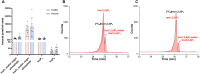
Inositol phosphates (InsPs) are ubiquitous in all eukaryotes. However, since there are 63 possible different phosphate ester isomers, the analysis of InsPs is challenging. In particular, InsP1, InsP2, and InsP3 already amass 41 different isomers, of which some occur as enantiomers. Profiling of these "lower" inositol phosphates in mammalian tissues requires powerful analytical methods and reference compounds. Here, we report an analysis of InsP2 and InsP3 with capillary electrophoresis coupled to electrospray ionization mass spectrometry (CE-ESI-MS). Using this method, the bacterial effector RipBL1 was analyzed and found to degrade InsP6 to Ins(1,2,3)P3, an understudied InsP3 isomer. This new reference molecule then aided us in the assignment of the isomeric identity of an InsP3 while profiling human samples: in urine and kidney stones, we describe for the first time the presence of defined and abundant InsP3 isomers, namely Ins(1,2,3)P3, Ins(1,2,6)P3 and/or Ins(2,3,4)P3.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Interaction of myoinositoltrisphosphate-phytase complex with the receptor for intercellular Ca2+ mobilization in plants.Biochemistry. 1996 Apr 16;35(15):4994-5001. doi: 10.1021/bi9525233. Biochemistry. 1996. PMID: 8664292
-
Formation of inositol polyphosphates in airway smooth muscle after muscarinic receptor stimulation.J Pharmacol Exp Ther. 1990 Feb;252(2):786-91. J Pharmacol Exp Ther. 1990. PMID: 2313600
-
Prostaglandin F2 alpha stimulates inositol 1,4,5-trisphosphate and inositol 1,3,4,5-tetrakisphosphate formation in bovine luteal cells.Endocrinology. 1991 Mar;128(3):1519-26. doi: 10.1210/endo-128-3-1519. Endocrinology. 1991. PMID: 1847860
-
Control of glomerulosa cell function by angiotensin II: transduction by G-proteins and inositol polyphosphates.Clin Exp Pharmacol Physiol. 1988 Jul;15(7):501-15. doi: 10.1111/j.1440-1681.1988.tb01108.x. Clin Exp Pharmacol Physiol. 1988. PMID: 3152162 Review.
-
Inositol Pyrophosphate Pathways and Mechanisms: What Can We Learn from Plants?Molecules. 2020 Jun 17;25(12):2789. doi: 10.3390/molecules25122789. Molecules. 2020. PMID: 32560343 Free PMC article. Review.
Cited by
-
Bacterial Pathogen Infection Triggers Magic Spot Nucleotide Signaling in Arabidopsis thaliana Chloroplasts through Specific RelA/SpoT Homologues.J Am Chem Soc. 2023 Jul 26;145(29):16081-16089. doi: 10.1021/jacs.3c04445. Epub 2023 Jul 12. J Am Chem Soc. 2023. PMID: 37437195 Free PMC article.
-
Biochemical and biophysical characterization of inositol-tetrakisphosphate 1-kinase inhibitors.J Biol Chem. 2025 Mar;301(3):108274. doi: 10.1016/j.jbc.2025.108274. Epub 2025 Feb 6. J Biol Chem. 2025. PMID: 39922495 Free PMC article.
-
Generation of inositol polyphosphates through a phospholipase C-independent pathway involving carbohydrate and sphingolipid metabolism in Trypanosoma cruzi.mBio. 2025 May 14;16(5):e0331824. doi: 10.1128/mbio.03318-24. Epub 2025 Apr 2. mBio. 2025. PMID: 40172212 Free PMC article.
-
CDS2 expression regulates de novo phosphatidic acid synthesis.Biochem J. 2024 Oct 16;481(20):1449-1473. doi: 10.1042/BCJ20240456. Biochem J. 2024. PMID: 39312194 Free PMC article.
-
Pools of Independently Cycling Inositol Phosphates Revealed by Pulse Labeling with 18O-Water.J Am Chem Soc. 2025 May 28;147(21):17626-17641. doi: 10.1021/jacs.4c16206. Epub 2025 May 15. J Am Chem Soc. 2025. PMID: 40372010 Free PMC article.
References
-
- Otto J. C. Kelly P. Chiou S. T. York J. D. Alterations in an inositol phosphate code through synergistic activation of a G protein and inositol phosphate kinases. Proc. Natl. Acad. Sci. U. S. A. 2007;104(40):15653–15658. doi: 10.1073/pnas.0705729104. doi: 10.1073/pnas.0705729104. - DOI - DOI - PMC - PubMed
-
- Dovey C. M. Diep J. Clarke B. P. Hale A. T. McNamara D. E. Guo H. Brown, Jr. N. W. Cao J. Y. Grace C. R. Gough P. J. Bertin J. Dixon S. J. Fiedler D. Mocarski E. S. Kaiser W. J. Moldoveanu T. York J. D. Carette J. E. MLKL Requires the Inositol Phosphate Code to Execute Necroptosis. Mol. Cell. 2018;70(5):936–948 e7. doi: 10.1016/j.molcel.2018.05.010. doi: 10.1016/j.molcel.2018.05.010. - DOI - DOI - PMC - PubMed
-
- Ito M. Fujii N. Wittwer C. Sasaki A. Tanaka M. Bittner T. Jessen H. J. Saiardi A. Takizawa S. Nagata E. Hydrophilic interaction liquid chromatography-tandem mass spectrometry for the quantitative analysis of mammalian-derived inositol poly/pyrophosphates. J. Chromatogr. A. 2018;1573:87–97. doi: 10.1016/j.chroma.2018.08.061. doi: 10.1016/j.chroma.2018.08.061. - DOI - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

