This is a preprint.
Multi-ancestry genome-wide study in >2.5 million individuals reveals heterogeneity in mechanistic pathways of type 2 diabetes and complications
Affiliations
- 1 Centre for Genetics and Genomics Versus Arthritis, Centre for Musculoskeletal Research, Division of Musculoskeletal and Dermatological Sciences, The University of Manchester, Manchester, UK.
- 2 Department of Diabetes and Metabolic Diseases, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan.
- 3 Department of Statistical Genetics, Osaka University Graduate School of Medicine, Suita, Japan.
- 4 Institute of Translational Genomics, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany.
- 5 Center for Precision Health Research, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA.
- 6 British Heart Foundation Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK.
- 7 Heart and Lung Research Institute, University of Cambridge, Cambridge, UK.
- 8 Department of Biostatistics and Center for Statistical Genetics, University of Michigan, Ann Arbor, MI, USA.
- 9 Department of Epidemiology, School of Public Health, Nanjing Medical University, Nanjing City, China.
- 10 Corporal Michael J Crescenz VA Medical Center, Philadelphia, PA, USA.
- 11 Department of Systems Pharmacology and Translational Therapeutics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 12 Department of Genetics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 13 Programs in Metabolism and Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA, USA.
- 14 Diabetes Unit and Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA, USA.
- 15 Consejo Nacional de Ciencia y Tecnología (CONACYT), Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
- 16 Department of Genome Informatics, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan.
- 17 Integrated Frontier Research for Medical Science Division, Institute for Open and Transdisciplinary Research Initiatives, Osaka University, Suita, Japan.
- 18 Laboratory for Systems Genetics, RIKEN Center for Integrative Medical Sciences, Kanagawa, Japan.
- 19 The Charles Bronfman Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
- 20 Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA.
- 21 Estonian Genome Centre, Institute of Genomics, University of Tartu, Tartu, Estonia.
- 22 Nuffield Department of Population Health, University of Oxford, Oxford, UK.
- 23 Medical Research Council Population Health Research Unit, University of Oxford, Oxford, UK.
- 24 The Institute for Translational Genomics and Population Sciences, Department of Pediatrics, The Lundquist Institute for Biomedical Innovation (formerly Los Angeles Biomedical Research Institute) at Harbor-UCLA Medical Center, Torrance, CA, USA.
- 25 Department of Epidemiology and Biostatistics, Imperial College London, London, UK.
- 26 Department of Cardiology, Ealing Hosptial, London NorthWest Healthcare NHS Trust, Middlesex, UK.
- 27 Division of Genome Science, Department of Precision Medicine, National Institute of Health, Cheongju-si, South Korea.
- 28 Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 29 Department of Gene Diagnostics and Therapeutics, Research Institute, National Center for Global Health and Medicine, Tokyo, Japan.
- 30 Institute of Epidemiology, Helmholtz Zentrum Munchen, German Research Center for Environmental Health, Neuherberg, Germany.
- 31 Department of Medicine, McMaster University, Hamilton, ON, Canada.
- 32 Population Health Research Institute, Hamilton Health Sciences and McMaster University, Hamilton, ON, Canada.
- 33 Public Health Informatics Unit, Department of Integrated Health Sciences, Nagoya University Graduate School of Medicine, Nagoya, Japan.
- 34 MRC Epidemiology Unit, Institute of Metabolic Science, University of Cambridge School of Clinical Medicine, Cambridge, UK.
- 35 Department of Health Data Science, University of Liverpool, Liverpool, UK.
- 36 Division of Translational Medicine and Human Genetics, University of Pennsylvania, Philadelphia, PA, USA.
- 37 Department of Epidemiology, Brown University School of Public Health, Providence, RI, USA.
- 38 Department of Anthropology, University of Toronto at Mississsauga, Mississauga, ON, Canada.
- 39 Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore, Singapore.
- 40 Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, MI, USA.
- 41 Center for Genomic Medicine, Kyoto University Graduate School of Medicine, Kyoto, Japan.
- 42 deCODE Genetics, Amgen Inc., Reykjavik, Iceland.
- 43 Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 44 Department of Biostatistics, Harvard University, Boston, MA, USA.
- 45 Division of Sleep and Circadian Disorders, Brigham and Women's Hospital, Boston, MA, USA.
- 46 Department of Medicine, Harvard University, Boston, MA, USA.
- 47 Institute of Genetic Epidemiology, Department of Data Driven Medicine, Faculty of Medicine and Medical Center, University of Freiburg, Freiburg, Germany.
- 48 Department of Epidemiology, Human Genetics, and Environmental Sciences, The University of Texas Health Science Center at Houston School of Public Health, Houston, TX, USA.
- 49 German Center for Diabetes Research (DZD), Neuherberg, Germany.
- 50 Research Unit of Molecular Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany.
- 51 Department of Population and Public Health Sciences, Keck School of Medicine of USC, Los Angeles, CA, USA.
- 52 Department of Cardiology, Leiden University Medical Center, Leiden, The Netherlands.
- 53 Section of Gerontology and Geriatrics, Department of Internal Medicine, Leiden University Medical Center, Leiden, The Netherlands.
- 54 Department of Internal Medicine, Seoul National University Hospital, Seoul, South Korea.
- 55 Division of Epidemiology, Department of Medicine, Institute for Medicine and Public Health, Vanderbilt Genetics Institute, Vanderbilt University Medical Center, Nashville, TN, USA.
- 56 Nuffield Department of Surgical Sciences, University of Oxford, Oxford, UK.
- 57 Institute for Population and Precision Health (IPPH), Biological Sciences Division, The University of Chicago, Chicago, IL, USA.
- 58 Department of Public Health Sciences and Center for Public Health Genomics, University of Virginia School of Medicine, Charlottesville, VA, USA.
- 59 Genomic Research on Complex Diseases (GRC-Group), CSIR-Centre for Cellular and Molecular Biology (CSIR-CCMB), Hyderabad, India.
- 60 Department of Medicine and Therapeutics, The Chinese University of Hong Kong, Hong Kong, China.
- 61 Chinese University of Hong Kong-Shanghai Jiao Tong University Joint Research Centre in Diabetes Genomics and Precision Medicine, The Chinese University of Hong Kong, Hong Kong, China.
- 62 Institute of Data Science, Korea University, Seoul, South Korea.
- 63 Division of Endocrinology, Metabolism, and Molecular Medicine, Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 64 Department of Health and Biomedical Informatics, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 65 Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan.
- 66 Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 67 Department of Human Genetics, Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, UK.
- 68 Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA.
- 69 Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
- 70 Center for Research on Genomics and Global Health, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA.
- 71 Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA, USA.
- 72 Division of Academics, Ochsner Health, New Orleans, LA, USA.
- 73 Division of Statistical Genomics, Washington University School of Medicine, St. Louis, MO, USA.
- 74 Department of Research and Evaluation, Division of Biostatistics Research, Kaiser Permanente of Southern California, Pasadena, CA, USA.
- 75 Department of Biomedical Science, Hallym University, Chuncheon, South Korea.
- 76 Harvard Medical School, Boston, MA, USA.
- 77 Metabolic Research Laboratories, Wellcome Trust-Medical Research Council Institute of Metabolic Science, Department of Clinical Biochemistry, University of Cambridge, Cambridge, UK.
- 78 Department of Epidemiology, University of Groningen, University Medical Centre Groningen, Groningen, The Netherlands.
- 79 Department of Bioinformatics, Isfahan University of Medical Sciences, Isfahan, Iran.
- 80 Department of Medicine, Division of Endocrinology, Diabetes and Metabolism, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
- 81 Department of Nutrition, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 82 Unidad de Investigación en Enfermedades Metabólicas and Departamento de Endocrinología y Metabolismo, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
- 83 Steno Diabetes Center Copenhagen, Herlev, Denmark.
- 84 The Bioinformatics Center, Department of Biology, University of Copenhagen, Copenhagen, Denmark.
- 85 Department of Health Research Methods, Evidence, and Impact, McMaster University, Hamilton, ON, Canada.
- 86 Department of Epidemiology and Prevention, Division of Public Health Sciences, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 87 Institute of Regional Health Research, University of Southern Denmark, Odense, Denmark.
- 88 Department of Clinical Biochemistry, Vejle Hospital, Vejle, Denmark.
- 89 Department of Medicine, Division of Endocrinology and Diabetes, Keck School of Medicine of USC, Los Angeles, CA, USA.
- 90 Department of Internal Medicine, University of Michigan, Ann Arbor, MI, USA.
- 91 British Heart Foundation Centre of Research Excellence, School of Clinical Medicine, Addenbrooke's Hospital, University of Cambridge, Cambridge, UK.
- 92 Health Data Research UK Cambridge, Wellcome Genome Campus and University of Cambridge, Hinxton, UK.
- 93 National Institute for Health and Care Research (NIHR) Blood and Transplant Unit (BTRU) in Donor Health and Behaviour, Heart and Lung Research Institute, University of Cambridge, Cambridge, UK.
- 94 Inserm U1283, CNRS UMR 8199, European Genomic Institute for Diabetes (EGID), Institut Pasteur de Lille, Lille University Hospital, Lille, France.
- 95 University of Lille, Lille, France.
- 96 Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong, China.
- 97 Hong Kong Institute of Diabetes and Obesity, The Chinese University of Hong Kong, Hong Kong, China.
- 98 Singapore Eye Research Institute, Singapore National Eye Centre, Singapore, Singapore.
- 99 Exeter Centre of Excellence in Diabetes (ExCEeD), Exeter Medical School, University of Exeter, Exeter, UK.
- 100 Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, UK.
- 101 Department of Biostatistics and Data Science, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 102 Division of Endocrinology and Metabolism, Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan.
- 103 Institute of Epidemiology and Preventive Medicine, National Taiwan University, Taipei, Taiwan.
- 104 Department of Medicine, University of Vermont, Colchester, VT, USA.
- 105 Section on Endocrinology and Metabolism, Department of Internal Medicine, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 106 Department of Medicine, Faculty of Medicine, University of Kelaniya, Ragama, Sri Lanka.
- 107 Department of Nutrition and Dietetics, Harokopio University of Athens, Athens, Greece.
- 108 Center for Genomics and Personalized Medicine Research, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 109 Carolina Population Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 110 Department of Nephrology and Medical Intensive Care Medicine, Charité Universitätsmedizin Berlin, Berlin, Germany.
- 111 Department of Nephrology and Hypertension, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany.
- 112 Department of Biostatistics, University of Washington, Seattle, WA, USA.
- 113 California Pacific Medical Center Research Institute, San Francisco, CA, USA.
- 114 Laboratory of Epidemiology and Population Sciences, National Institute on Aging, National Institutes of Health, Baltimore, MD, USA.
- 115 Institute of Mathematics and Statistics, University of Tartu, Tartu, Estonia.
- 116 Robertson Centre for Biostatistics, University of Glasgow, Glasgow, UK.
- 117 Department of Epidemiology, Erasmus MC University Medical Center, Rotterdam, The Netherlands.
- 118 Genetics of Complex Traits, University of Exeter Medical School, University of Exeter, Exeter, UK.
- 119 Department of Internal Medicine, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 120 Department of Medicine, Division of Endocrinology and Metabolism, Lundquist Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA.
- 121 Department of Public Health and Caring Sciences, Uppsala University, Uppsala, Sweden.
- 122 Centro de Estudios en Diabetes, Unidad de Investigacion en Diabetes y Riesgo Cardiovascular, Centro de Investigacion en Salud Poblacional, Instituto Nacional de Salud Publica, Mexico City, Mexico.
- 123 Department of Laboratory Medicine and Pathology, University of Minnesota, Minneapolis, MN, USA.
- 124 Genomics and Computational Biology Graduate Group, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 125 University of Exeter Medical School, University of Exeter, Exeter, UK.
- 126 Institute for Clinical Diabetology, German Diabetes Center, Leibniz Center for Diabetes Research at Heinrich Heine University Düsseldorf, Düsseldorf, Germany.
- 127 Department of Endocrinology and Diabetology, Medical Faculty and University Hospital Dusseldorf, Heinrich Heine University Düsseldorf, Düsseldorf, Germany.
- 128 Laboratory for Genomics of Diabetes and Metabolism, RIKEN Center for Integrative Medical Sciences, Kanagawa, Japan.
- 129 Department of Biostatistics, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 130 Department of Internal Medicine, Diabetes and Metabolism Research Center, The Ohio State University Wexner Medical Center, Columbus, OH, USA.
- 131 Center for Global Cardiometabolic Health, Brown University, Providence, RI, USA.
- 132 Shanghai-MOST Key Laboratory of Health and Disease Genomics, Chinese National Human Genome Center at Shanghai (CHGC) and Shanghai Institute for Biomedical and Pharmaceutical Technologies (SIBPT), Shanghai, China.
- 133 Division of Endocrine and Metabolism, Tri-Service General Hospital Songshan Branch, Taipei, Taiwan.
- 134 School of Medicine, National Defense Medical Center, Taipei, Taiwan.
- 135 Division of Genome Science, Department of Precision Medicine, National Institute of Health, Cheongju-si, Korea.
- 136 Section of Endocrinology and Metabolism, Department of Medicine, Taipei Veterans General Hospital, Taipei, Taiwan.
- 137 School of Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan.
- 138 Department of Environmental and Preventive Medicine, Jichi Medical University School of Medicine, Shimotsuke, Japan.
- 139 University of Chicago Research Bangladesh, Dhaka, Bangladesh.
- 140 Institute of Molecular and Clinical Ophthalmology Basel, Basel, Switzerland.
- 141 Center for Clinical Research and Prevention, Bispebjerg and Frederiksberg Hospital, Frederiksberg, Denmark.
- 142 Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 143 Faculty of Medicine, Aalborg University, Aalborg, Denmark.
- 144 Department of Clinical Diabetes, Endocrinology and Metabolism, Department of Translational Research and Cellular Therapeutics, City of Hope, Duarte, CA, USA.
- 145 Department of Public Health, Faculty of Medicine, University of Kelaniya, Ragama, Sri Lanka.
- 146 Department of Clinical Gene Therapy, Osaka University Graduate School of Medicine, Osaka, Japan.
- 147 Department of Geriatric and General Medicine, Graduate School of Medicine, Osaka University, Osaka, Japan.
- 148 Division of General Internal Medicine and Geriatrics, Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 149 Center for Health Information Partnerships, Institute for Public Health and Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 150 Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore, Singapore.
- 151 Department of Molecular Cell Biology, Sungkyunkwan University School of Medicine, Suwon, South Korea.
- 152 Institute of Genetic Epidemiology, Medical University of Innsbruck, Innsbruck, Austria.
- 153 Institute of Clinical Medicine, Internal Medicine, University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland.
- 154 Department of Medicine, University of Colorado Denver, Anschutz Medical Campus, Aurora, CO, USA.
- 155 VA Salt Lake City Health Care System, Salt Lake City, UT, USA.
- 156 Department of Internal Medicine, University of Utah School of Medicine, Salt Lake City, UT, USA.
- 157 Severance Biomedical Science Institute and Department of Internal Medicine, Yonsei University College of Medicine, Seoul, South Korea.
- 158 Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, South Korea.
- 159 USC-Office of Population Studies Foundation Inc., University of San Carlos, Cebu City, Philippines.
- 160 Department of Medicine, Harvard Medical School, Boston, MA, USA.
- 161 Division of General Internal Medicine, Massachusetts General Hospital, Boston, MA, USA.
- 162 Department of Epidemiology and Biostatistics, School of Public Health, Peking University, Beijing, China.
- 163 Peking University Center for Public Health and Epidemic Preparedness and Response, Beijing, China.
- 164 Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, The Netherlands.
- 165 Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, UK.
- 166 Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA.
- 167 Big Data Institute, Li Ka Shing Centre For Health Information and Discovery, University of Oxford, Oxford, UK.
- 168 Department of Clinical Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 169 Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA.
- 170 Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore and National University Health System, Singapore, Singapore.
- 171 McDonnell Genome Institute, Washington University School of Medicine, St Louis, MO, USA.
- 172 Department of Medicine, Division of Genomics and Bioinformatics, Washington University School of Medicine, St Louis, MO, USA.
- 173 Present address: Regeneron Genetics Center, Tarrytown, NY, USA.
- 174 Department of Biostatistics and Data Science, The University of Texas Health Science Center at Houston School of Public Health, Houston, TX, USA.
- 175 Department of Clinical Sciences, Diabetes and Endocrinology, Lund University Diabetes Centre, Malmö, Sweden.
- 176 Department of Clinical Science, Center for Diabetes Research, University of Bergen, Bergen, Norway.
- 177 Department of Advanced Genomic and Laboratory Medicine, Graduate School of Medicine, University of the Ryukyus, Okinawa, Japan.
- 178 Division of Clinical Laboratory and Blood Transfusion, University of the Ryukyus Hospital, Okinawa, Japan.
- 179 Dromokaiteio Psychiatric Hospital, National and Kapodistrian University of Athens, Athens, Greece.
- 180 Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Tokyo, Japan.
- 181 Institute of Human Genetics, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany.
- 182 Institute of Human Genetics, Technical University Munich, Munich, Germany.
- 183 German Centre for Cardiovascular Research (DZHK), Partner Site Munich Heart Alliance, Munich, Germany.
- 184 The Usher Institute to the Population Health Sciences and Informatics, University of Edinburgh, Edinburgh, UK.
- 185 Department of Medicine and Pharmacology, New York Medical College, Valhalla, NY, USA.
- 186 Data Tecnica International LLC, Glen Echo, MD, USA.
- 187 Center for Alzheimer's and Related Dementias, National Institutes of Health, Bethesda, MD, USA.
- 188 William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London, UK.
- 189 Laboratory of Statistical Immunology, Immunology Frontier Research Center (WPI-IFReC), Osaka University, Suita, Japan.
- 190 Instituto Nacional de Medicina Genómica, Mexico City, Mexico.
- 191 Division of Pulmonary, Allergy, and Critical Care Medicine, Department of Medicine, University of Pittsburgh, Pittsburgh, PA, USA.
- 192 Division of Epidemiology and Community Health, School of Public Health, University of Minnesota, Minneapolis, MN, USA.
- 193 Institute for Medical Information Processing, Biometry and Epidemiology, Ludwig Maximilians Universität München, Munich, Germany.
- 194 Department of Diabetes and Endocrinology, Nelson R Mandela School of Medicine, College of Health Sciences, University of KwaZulu-Natal, Durban, South Africa.
- 195 Academy of Scientific and Innovative Research, CSIR-Human Resource Development Campus, Ghaziabad, India.
- 196 Genomics and Molecular Medicine Unit, CSIR-Institute of Genomics and Integrative Biology, New Delhi, India.
- 197 Department of Preventive Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 198 Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
- 199 Ophthalmology and Visual Sciences Academic Clinical Program (Eye ACP), Duke-NUS Medical School, Singapore, Singapore.
- 200 Department of Ophthalmology, Yong Loo Lin School of Medicine, National University of Singapore and National University Health System, Singapore, Singapore.
- 201 Institute of Cardiovascular and Medical Sciences, University of Glasgow, Glasgow, UK.
- 202 Survey Research Center, Institute for Social Research, University of Michigan, Ann Arbor, MI, USA.
- 203 Institute of Genetic Epidemiology, Helmholtz Zentrum Munchen, German Research Center for Environmental Health, Neuherberg, Germany.
- 204 Institute for Medical Biostatistics, Epidemiology and Informatics (IMBEI), University Medical Center, Johannes Gutenberg University, Mainz, Germany.
- 205 Chair of Genetic Epidemiology, Institute of Medical Information Processing, Biometry, and Epidemiology, Faculty of Medicine, Ludwig Maximilians Universität München, Munich, Germany.
- 206 Faculty of Medicine, University of Iceland, Reykjavik, Iceland.
- 207 Faculty of Medicine, Macau University of Science and Technology, Macau, China.
- 208 Department of Medical Genetics and Medical Research, China Medical University Hospital, Taichung, Taiwan.
- 209 Population Health Unit, Finnish Institute for Health and Welfare, Helsinki, Finland, Finnish Institute for Health and Welfare, Helsinki, Finland.
- 210 National School of Public Health, Madrid, Spain.
- 211 Department of Public Health, University of Helsinki, Helsinki, Finland.
- 212 Diabetes Research Group, King Abdulaziz University, Jeddah, Saudi Arabia.
- 213 Unidad de Biología Molecular y Medicina Genómica, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
- 214 Departamento de Medicina Genómica y Toxiología Ambiental, Instituto de Investigaciones Biomédicas, UNAM, Mexico City, Mexico.
- 215 Unidad de Investigacion Medica en Bioquimica, Hospital de Especialidades, Centro Medico Nacional Siglo XXI, Instituto Mexicano del Seguro Social, Mexico City, Mexico.
- 216 Einthoven Laboratory for Experimental Vascular Medicine, Leiden University Medical Center, Leiden, The Netherlands.
- 217 Department of Human Genetics, Leiden University Medical Center, Leiden, The Netherlands.
- 218 Department of Clinical Chemistry, Laboratory of Genetic Metabolic Disease, Amsterdam University Medical Center, Amsterdam, The Netherlands.
- 219 Southern California Eye Institute, CHA Hollywood Presbyterian Hospital, Los Angeles, CA, USA.
- 220 Unidad de Investigación Médica en Epidemiologia Clinica, Hospital de Especialidades, Centro Medico Nacional Siglo XXI, Instituto Mexicano del Seguro Social, Mexico City, Mexico.
- 221 Department of Internal Medicine, Division of Endocrinology, Leiden University Medical Center, Leiden, The Netherlands.
- 222 Department of Public Health, Aarhus University, Aarhus, Denmark.
- 223 Danish Diabetes Academy, Odense, Denmark.
- 224 Diabetology Research Centre, King Edward Memorial Hospital and Research Centre, Pune, India.
- 225 Department of Medical Biochemistry, Kurume University School of Medicine, Kurume, Japan.
- 226 Department of Pediatrics, Osaka University Graduate School of Medicine, Suita, Japan.
- 227 Division of Cancer Control and Population Sciences, UPMC Hillman Cancer Center, University of Pittsburgh, Pittsburgh, PA, USA.
- 228 Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA.
- 229 Department of Pediatrics, Division of Genetic and Genomic Medicine, UCI Irvine School of Medicine, Irvine, CA, USA.
- 230 Department of Anti-Aging Medicine, Ehime University Graduate School of Medicine, Ehime, Japan.
- 231 Division of Pulmonary, Critical Care, and Sleep Medicine, Beth Israel Deaconess Medical Center, Boston, MA, USA.
- 232 Department of Medical Sciences, Uppsala University, Uppsala, Sweden.
- 233 Institute of Molecular Medicine, The University of Texas Health Science Center at Houston School of Public Health, Houston, TX, USA.
- 234 Human Genetics Center, University of Texas Health Science Center at Houston, Houston, TX, US.
- 235 Department of Medicine, Stanford University School of Medicine, Stanford, CA, USA.
- 236 Department of Medical Sciences, Molecular Epidemiology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden.
- 237 Department of Epidemiology, University of Washington, Seattle, WA, USA.
- 238 Department of Health Systems and Population Health, University of Washington, Seattle, WA, USA.
- 239 Beijing Institute of Ophthalmology, Ophthalmology and Visual Sciences Key Laboratory, Beijing Tongren Hospital, Capital Medical University, Beijing, China.
- 240 Department of Medicine, Division of Cardiology, Duke University School of Medicine, Durham, NC, USA.
- 241 Kurume University School of Medicine, Kurume, Japan.
- 242 Department of Medicine, McGill University, Montreal, QC, Canada.
- 243 Department of Human Genetics, McGill University, Montreal, QC, Canada.
- 244 Department of Metabolism, Digestion and Reproduction, Imperial College London, London, UK.
- 245 Department of Physiology and Biophysics, University of Mississippi Medical Center, Jackson, MS, USA.
- 246 Division of Endocrinology and Metabolism, Department of Medicine, Taichung Veterans General Hospital, Taichung, Taiwan.
- 247 Center for Genetic Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 248 Department of Anthropology, Northwestern University, Evanston, IL, USA.
- 249 Department of Endocrinology, Helsinki University Hospital, Helsinki, Finland.
- 250 Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland.
- 251 Folkhalsan Research Center, Helsinki, Finland.
- 252 Lund University Diabetes Centre, Malmö, Sweden.
- 253 Systems Genomics Laboratory, School of Biotechnology, Jawaharlal Nehru University, New Delhi, India.
- 254 Department of Pathology and Molecular Medicine, McMaster University, Hamilton, ON, Canada.
- 255 Deceased.
- 256 Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, South Korea.
- 257 Netherlands Heart Institute, Utrecht, The Netherlands.
- 258 Center for Public Health Genomics, University of Virginia School of Medicine, Charlottesville, VA, USA.
- 259 Research Unit of Molecular Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Munich, Germany.
- 260 Duke-NUS Medical School, Singapore, Singapore.
- 261 Department of Epidemiology, Biostatistics and Occupational Health, McGill University, Montreal, QC, Canada.
- 262 Singapore Institute for Clinical Sciences, Agency for Science Technology and Research (A*STAR), Singapore, Singapore.
- 263 Healthy Longevity Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
- 264 Center for Diabetes Research, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 265 Department of Biochemistry, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 266 Pat Macpherson Centre for Pharmacogenetics and Pharmacogenomics, University of Dundee, Dundee, UK.
- 267 Imperial College Healthcare NHS Trust, Imperial College London, London, UK.
- 268 MRC-PHE Centre for Environment and Health, Imperial College London, London, UK.
- 269 National Heart and Lung Institute, Imperial College London, London, UK.
- 270 Department of Medicine, Brown University Alpert School of Medicine, Providence, RI, USA.
- 271 Department of Medicine, Columbia University Irving Medical Center, New York, NY, USA.
- 272 Department of Cardiology, Columbia University Irving Medical Center, New York, NY, USA.
- 273 Center for Non-Communicable Diseases, Karachi, Pakistan.
- 274 Computational Medicine, Berlin Institute of Health at Charité Universitätsmedizin, Berlin, Germany.
- 275 Precision Healthcare University Research Institute, Queen Mary University of London, London, UK.
- 276 Department of Preventive Medicine, Keck School of Medicine of USC, Los Angeles, CA, USA.
- 277 The Mindich Child Health and Development Institute, Ichan School of Medicine at Mount Sinai, New York, NY, USA.
- 278 Division of Translational Medicine and Therapeutics, Department of Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 279 Institute for Translational Medicine and Therapeutics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 280 Department of Pediatrics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 281 Center for Precision Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 282 Institute for Biomedical Informatics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 283 Department of Psychiatry, University of Michigan, Ann Arbor, MI, USA.
- 284 All of Us Research Program, National Institutes of Health, Bethesda, MD, USA.
- 285 Toranomon Hospital, Tokyo, Japan.
- 286 Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore.
- 287 Vanderbilt Genetics Institute, Division of Genetic Medicine, Vanderbilt University Medical Center, Nashville, TN, USA.
- 288 VA Palo Alto Health Care System, Palo Alto, CA, USA.
- 289 Stanford Cardiovascular Institute, Stanford University School of Medicine, Stanford, CA, USA.
- 290 Department of Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 291 Oxford Centre for Diabetes, Endocrinology and Metabolism, Radcliffe Department of Medicine, University of Oxford, Oxford, UK.
- 292 Oxford NIHR Biomedical Research Centre, Churchill Hosptial, Oxford University Hospitals NHS Foundation Trust, Oxford, UK.
- 293 Present address: Genentech, South San Francisco, CA, USA.
- 294 Department of Biostatistics and Epidemiology, University of Massachusetts Amherst, Amherst, MA, USA.
- 295 Division of Epidemiology, Department of Biostatistics, Epidemiology and Informatics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 296 TUM School of Medicine, Technical University of Munich and Klinikum Rechts der Isar, Munich, Germany.
- PMID: 37034649
- PMCID: PMC10081410
- DOI: 10.1101/2023.03.31.23287839
Multi-ancestry genome-wide study in >2.5 million individuals reveals heterogeneity in mechanistic pathways of type 2 diabetes and complications
Authors
Affiliations
- 1 Centre for Genetics and Genomics Versus Arthritis, Centre for Musculoskeletal Research, Division of Musculoskeletal and Dermatological Sciences, The University of Manchester, Manchester, UK.
- 2 Department of Diabetes and Metabolic Diseases, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan.
- 3 Department of Statistical Genetics, Osaka University Graduate School of Medicine, Suita, Japan.
- 4 Institute of Translational Genomics, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany.
- 5 Center for Precision Health Research, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA.
- 6 British Heart Foundation Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK.
- 7 Heart and Lung Research Institute, University of Cambridge, Cambridge, UK.
- 8 Department of Biostatistics and Center for Statistical Genetics, University of Michigan, Ann Arbor, MI, USA.
- 9 Department of Epidemiology, School of Public Health, Nanjing Medical University, Nanjing City, China.
- 10 Corporal Michael J Crescenz VA Medical Center, Philadelphia, PA, USA.
- 11 Department of Systems Pharmacology and Translational Therapeutics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 12 Department of Genetics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 13 Programs in Metabolism and Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA, USA.
- 14 Diabetes Unit and Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA, USA.
- 15 Consejo Nacional de Ciencia y Tecnología (CONACYT), Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
- 16 Department of Genome Informatics, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan.
- 17 Integrated Frontier Research for Medical Science Division, Institute for Open and Transdisciplinary Research Initiatives, Osaka University, Suita, Japan.
- 18 Laboratory for Systems Genetics, RIKEN Center for Integrative Medical Sciences, Kanagawa, Japan.
- 19 The Charles Bronfman Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
- 20 Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA.
- 21 Estonian Genome Centre, Institute of Genomics, University of Tartu, Tartu, Estonia.
- 22 Nuffield Department of Population Health, University of Oxford, Oxford, UK.
- 23 Medical Research Council Population Health Research Unit, University of Oxford, Oxford, UK.
- 24 The Institute for Translational Genomics and Population Sciences, Department of Pediatrics, The Lundquist Institute for Biomedical Innovation (formerly Los Angeles Biomedical Research Institute) at Harbor-UCLA Medical Center, Torrance, CA, USA.
- 25 Department of Epidemiology and Biostatistics, Imperial College London, London, UK.
- 26 Department of Cardiology, Ealing Hosptial, London NorthWest Healthcare NHS Trust, Middlesex, UK.
- 27 Division of Genome Science, Department of Precision Medicine, National Institute of Health, Cheongju-si, South Korea.
- 28 Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 29 Department of Gene Diagnostics and Therapeutics, Research Institute, National Center for Global Health and Medicine, Tokyo, Japan.
- 30 Institute of Epidemiology, Helmholtz Zentrum Munchen, German Research Center for Environmental Health, Neuherberg, Germany.
- 31 Department of Medicine, McMaster University, Hamilton, ON, Canada.
- 32 Population Health Research Institute, Hamilton Health Sciences and McMaster University, Hamilton, ON, Canada.
- 33 Public Health Informatics Unit, Department of Integrated Health Sciences, Nagoya University Graduate School of Medicine, Nagoya, Japan.
- 34 MRC Epidemiology Unit, Institute of Metabolic Science, University of Cambridge School of Clinical Medicine, Cambridge, UK.
- 35 Department of Health Data Science, University of Liverpool, Liverpool, UK.
- 36 Division of Translational Medicine and Human Genetics, University of Pennsylvania, Philadelphia, PA, USA.
- 37 Department of Epidemiology, Brown University School of Public Health, Providence, RI, USA.
- 38 Department of Anthropology, University of Toronto at Mississsauga, Mississauga, ON, Canada.
- 39 Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore, Singapore.
- 40 Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, MI, USA.
- 41 Center for Genomic Medicine, Kyoto University Graduate School of Medicine, Kyoto, Japan.
- 42 deCODE Genetics, Amgen Inc., Reykjavik, Iceland.
- 43 Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 44 Department of Biostatistics, Harvard University, Boston, MA, USA.
- 45 Division of Sleep and Circadian Disorders, Brigham and Women's Hospital, Boston, MA, USA.
- 46 Department of Medicine, Harvard University, Boston, MA, USA.
- 47 Institute of Genetic Epidemiology, Department of Data Driven Medicine, Faculty of Medicine and Medical Center, University of Freiburg, Freiburg, Germany.
- 48 Department of Epidemiology, Human Genetics, and Environmental Sciences, The University of Texas Health Science Center at Houston School of Public Health, Houston, TX, USA.
- 49 German Center for Diabetes Research (DZD), Neuherberg, Germany.
- 50 Research Unit of Molecular Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany.
- 51 Department of Population and Public Health Sciences, Keck School of Medicine of USC, Los Angeles, CA, USA.
- 52 Department of Cardiology, Leiden University Medical Center, Leiden, The Netherlands.
- 53 Section of Gerontology and Geriatrics, Department of Internal Medicine, Leiden University Medical Center, Leiden, The Netherlands.
- 54 Department of Internal Medicine, Seoul National University Hospital, Seoul, South Korea.
- 55 Division of Epidemiology, Department of Medicine, Institute for Medicine and Public Health, Vanderbilt Genetics Institute, Vanderbilt University Medical Center, Nashville, TN, USA.
- 56 Nuffield Department of Surgical Sciences, University of Oxford, Oxford, UK.
- 57 Institute for Population and Precision Health (IPPH), Biological Sciences Division, The University of Chicago, Chicago, IL, USA.
- 58 Department of Public Health Sciences and Center for Public Health Genomics, University of Virginia School of Medicine, Charlottesville, VA, USA.
- 59 Genomic Research on Complex Diseases (GRC-Group), CSIR-Centre for Cellular and Molecular Biology (CSIR-CCMB), Hyderabad, India.
- 60 Department of Medicine and Therapeutics, The Chinese University of Hong Kong, Hong Kong, China.
- 61 Chinese University of Hong Kong-Shanghai Jiao Tong University Joint Research Centre in Diabetes Genomics and Precision Medicine, The Chinese University of Hong Kong, Hong Kong, China.
- 62 Institute of Data Science, Korea University, Seoul, South Korea.
- 63 Division of Endocrinology, Metabolism, and Molecular Medicine, Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 64 Department of Health and Biomedical Informatics, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 65 Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan.
- 66 Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 67 Department of Human Genetics, Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, UK.
- 68 Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA.
- 69 Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
- 70 Center for Research on Genomics and Global Health, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA.
- 71 Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA, USA.
- 72 Division of Academics, Ochsner Health, New Orleans, LA, USA.
- 73 Division of Statistical Genomics, Washington University School of Medicine, St. Louis, MO, USA.
- 74 Department of Research and Evaluation, Division of Biostatistics Research, Kaiser Permanente of Southern California, Pasadena, CA, USA.
- 75 Department of Biomedical Science, Hallym University, Chuncheon, South Korea.
- 76 Harvard Medical School, Boston, MA, USA.
- 77 Metabolic Research Laboratories, Wellcome Trust-Medical Research Council Institute of Metabolic Science, Department of Clinical Biochemistry, University of Cambridge, Cambridge, UK.
- 78 Department of Epidemiology, University of Groningen, University Medical Centre Groningen, Groningen, The Netherlands.
- 79 Department of Bioinformatics, Isfahan University of Medical Sciences, Isfahan, Iran.
- 80 Department of Medicine, Division of Endocrinology, Diabetes and Metabolism, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
- 81 Department of Nutrition, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 82 Unidad de Investigación en Enfermedades Metabólicas and Departamento de Endocrinología y Metabolismo, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
- 83 Steno Diabetes Center Copenhagen, Herlev, Denmark.
- 84 The Bioinformatics Center, Department of Biology, University of Copenhagen, Copenhagen, Denmark.
- 85 Department of Health Research Methods, Evidence, and Impact, McMaster University, Hamilton, ON, Canada.
- 86 Department of Epidemiology and Prevention, Division of Public Health Sciences, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 87 Institute of Regional Health Research, University of Southern Denmark, Odense, Denmark.
- 88 Department of Clinical Biochemistry, Vejle Hospital, Vejle, Denmark.
- 89 Department of Medicine, Division of Endocrinology and Diabetes, Keck School of Medicine of USC, Los Angeles, CA, USA.
- 90 Department of Internal Medicine, University of Michigan, Ann Arbor, MI, USA.
- 91 British Heart Foundation Centre of Research Excellence, School of Clinical Medicine, Addenbrooke's Hospital, University of Cambridge, Cambridge, UK.
- 92 Health Data Research UK Cambridge, Wellcome Genome Campus and University of Cambridge, Hinxton, UK.
- 93 National Institute for Health and Care Research (NIHR) Blood and Transplant Unit (BTRU) in Donor Health and Behaviour, Heart and Lung Research Institute, University of Cambridge, Cambridge, UK.
- 94 Inserm U1283, CNRS UMR 8199, European Genomic Institute for Diabetes (EGID), Institut Pasteur de Lille, Lille University Hospital, Lille, France.
- 95 University of Lille, Lille, France.
- 96 Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong, China.
- 97 Hong Kong Institute of Diabetes and Obesity, The Chinese University of Hong Kong, Hong Kong, China.
- 98 Singapore Eye Research Institute, Singapore National Eye Centre, Singapore, Singapore.
- 99 Exeter Centre of Excellence in Diabetes (ExCEeD), Exeter Medical School, University of Exeter, Exeter, UK.
- 100 Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, UK.
- 101 Department of Biostatistics and Data Science, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 102 Division of Endocrinology and Metabolism, Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan.
- 103 Institute of Epidemiology and Preventive Medicine, National Taiwan University, Taipei, Taiwan.
- 104 Department of Medicine, University of Vermont, Colchester, VT, USA.
- 105 Section on Endocrinology and Metabolism, Department of Internal Medicine, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 106 Department of Medicine, Faculty of Medicine, University of Kelaniya, Ragama, Sri Lanka.
- 107 Department of Nutrition and Dietetics, Harokopio University of Athens, Athens, Greece.
- 108 Center for Genomics and Personalized Medicine Research, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 109 Carolina Population Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 110 Department of Nephrology and Medical Intensive Care Medicine, Charité Universitätsmedizin Berlin, Berlin, Germany.
- 111 Department of Nephrology and Hypertension, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany.
- 112 Department of Biostatistics, University of Washington, Seattle, WA, USA.
- 113 California Pacific Medical Center Research Institute, San Francisco, CA, USA.
- 114 Laboratory of Epidemiology and Population Sciences, National Institute on Aging, National Institutes of Health, Baltimore, MD, USA.
- 115 Institute of Mathematics and Statistics, University of Tartu, Tartu, Estonia.
- 116 Robertson Centre for Biostatistics, University of Glasgow, Glasgow, UK.
- 117 Department of Epidemiology, Erasmus MC University Medical Center, Rotterdam, The Netherlands.
- 118 Genetics of Complex Traits, University of Exeter Medical School, University of Exeter, Exeter, UK.
- 119 Department of Internal Medicine, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 120 Department of Medicine, Division of Endocrinology and Metabolism, Lundquist Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA.
- 121 Department of Public Health and Caring Sciences, Uppsala University, Uppsala, Sweden.
- 122 Centro de Estudios en Diabetes, Unidad de Investigacion en Diabetes y Riesgo Cardiovascular, Centro de Investigacion en Salud Poblacional, Instituto Nacional de Salud Publica, Mexico City, Mexico.
- 123 Department of Laboratory Medicine and Pathology, University of Minnesota, Minneapolis, MN, USA.
- 124 Genomics and Computational Biology Graduate Group, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 125 University of Exeter Medical School, University of Exeter, Exeter, UK.
- 126 Institute for Clinical Diabetology, German Diabetes Center, Leibniz Center for Diabetes Research at Heinrich Heine University Düsseldorf, Düsseldorf, Germany.
- 127 Department of Endocrinology and Diabetology, Medical Faculty and University Hospital Dusseldorf, Heinrich Heine University Düsseldorf, Düsseldorf, Germany.
- 128 Laboratory for Genomics of Diabetes and Metabolism, RIKEN Center for Integrative Medical Sciences, Kanagawa, Japan.
- 129 Department of Biostatistics, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 130 Department of Internal Medicine, Diabetes and Metabolism Research Center, The Ohio State University Wexner Medical Center, Columbus, OH, USA.
- 131 Center for Global Cardiometabolic Health, Brown University, Providence, RI, USA.
- 132 Shanghai-MOST Key Laboratory of Health and Disease Genomics, Chinese National Human Genome Center at Shanghai (CHGC) and Shanghai Institute for Biomedical and Pharmaceutical Technologies (SIBPT), Shanghai, China.
- 133 Division of Endocrine and Metabolism, Tri-Service General Hospital Songshan Branch, Taipei, Taiwan.
- 134 School of Medicine, National Defense Medical Center, Taipei, Taiwan.
- 135 Division of Genome Science, Department of Precision Medicine, National Institute of Health, Cheongju-si, Korea.
- 136 Section of Endocrinology and Metabolism, Department of Medicine, Taipei Veterans General Hospital, Taipei, Taiwan.
- 137 School of Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan.
- 138 Department of Environmental and Preventive Medicine, Jichi Medical University School of Medicine, Shimotsuke, Japan.
- 139 University of Chicago Research Bangladesh, Dhaka, Bangladesh.
- 140 Institute of Molecular and Clinical Ophthalmology Basel, Basel, Switzerland.
- 141 Center for Clinical Research and Prevention, Bispebjerg and Frederiksberg Hospital, Frederiksberg, Denmark.
- 142 Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 143 Faculty of Medicine, Aalborg University, Aalborg, Denmark.
- 144 Department of Clinical Diabetes, Endocrinology and Metabolism, Department of Translational Research and Cellular Therapeutics, City of Hope, Duarte, CA, USA.
- 145 Department of Public Health, Faculty of Medicine, University of Kelaniya, Ragama, Sri Lanka.
- 146 Department of Clinical Gene Therapy, Osaka University Graduate School of Medicine, Osaka, Japan.
- 147 Department of Geriatric and General Medicine, Graduate School of Medicine, Osaka University, Osaka, Japan.
- 148 Division of General Internal Medicine and Geriatrics, Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 149 Center for Health Information Partnerships, Institute for Public Health and Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 150 Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore, Singapore.
- 151 Department of Molecular Cell Biology, Sungkyunkwan University School of Medicine, Suwon, South Korea.
- 152 Institute of Genetic Epidemiology, Medical University of Innsbruck, Innsbruck, Austria.
- 153 Institute of Clinical Medicine, Internal Medicine, University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland.
- 154 Department of Medicine, University of Colorado Denver, Anschutz Medical Campus, Aurora, CO, USA.
- 155 VA Salt Lake City Health Care System, Salt Lake City, UT, USA.
- 156 Department of Internal Medicine, University of Utah School of Medicine, Salt Lake City, UT, USA.
- 157 Severance Biomedical Science Institute and Department of Internal Medicine, Yonsei University College of Medicine, Seoul, South Korea.
- 158 Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, South Korea.
- 159 USC-Office of Population Studies Foundation Inc., University of San Carlos, Cebu City, Philippines.
- 160 Department of Medicine, Harvard Medical School, Boston, MA, USA.
- 161 Division of General Internal Medicine, Massachusetts General Hospital, Boston, MA, USA.
- 162 Department of Epidemiology and Biostatistics, School of Public Health, Peking University, Beijing, China.
- 163 Peking University Center for Public Health and Epidemic Preparedness and Response, Beijing, China.
- 164 Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, The Netherlands.
- 165 Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, UK.
- 166 Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA.
- 167 Big Data Institute, Li Ka Shing Centre For Health Information and Discovery, University of Oxford, Oxford, UK.
- 168 Department of Clinical Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 169 Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA.
- 170 Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore and National University Health System, Singapore, Singapore.
- 171 McDonnell Genome Institute, Washington University School of Medicine, St Louis, MO, USA.
- 172 Department of Medicine, Division of Genomics and Bioinformatics, Washington University School of Medicine, St Louis, MO, USA.
- 173 Present address: Regeneron Genetics Center, Tarrytown, NY, USA.
- 174 Department of Biostatistics and Data Science, The University of Texas Health Science Center at Houston School of Public Health, Houston, TX, USA.
- 175 Department of Clinical Sciences, Diabetes and Endocrinology, Lund University Diabetes Centre, Malmö, Sweden.
- 176 Department of Clinical Science, Center for Diabetes Research, University of Bergen, Bergen, Norway.
- 177 Department of Advanced Genomic and Laboratory Medicine, Graduate School of Medicine, University of the Ryukyus, Okinawa, Japan.
- 178 Division of Clinical Laboratory and Blood Transfusion, University of the Ryukyus Hospital, Okinawa, Japan.
- 179 Dromokaiteio Psychiatric Hospital, National and Kapodistrian University of Athens, Athens, Greece.
- 180 Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Tokyo, Japan.
- 181 Institute of Human Genetics, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany.
- 182 Institute of Human Genetics, Technical University Munich, Munich, Germany.
- 183 German Centre for Cardiovascular Research (DZHK), Partner Site Munich Heart Alliance, Munich, Germany.
- 184 The Usher Institute to the Population Health Sciences and Informatics, University of Edinburgh, Edinburgh, UK.
- 185 Department of Medicine and Pharmacology, New York Medical College, Valhalla, NY, USA.
- 186 Data Tecnica International LLC, Glen Echo, MD, USA.
- 187 Center for Alzheimer's and Related Dementias, National Institutes of Health, Bethesda, MD, USA.
- 188 William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London, UK.
- 189 Laboratory of Statistical Immunology, Immunology Frontier Research Center (WPI-IFReC), Osaka University, Suita, Japan.
- 190 Instituto Nacional de Medicina Genómica, Mexico City, Mexico.
- 191 Division of Pulmonary, Allergy, and Critical Care Medicine, Department of Medicine, University of Pittsburgh, Pittsburgh, PA, USA.
- 192 Division of Epidemiology and Community Health, School of Public Health, University of Minnesota, Minneapolis, MN, USA.
- 193 Institute for Medical Information Processing, Biometry and Epidemiology, Ludwig Maximilians Universität München, Munich, Germany.
- 194 Department of Diabetes and Endocrinology, Nelson R Mandela School of Medicine, College of Health Sciences, University of KwaZulu-Natal, Durban, South Africa.
- 195 Academy of Scientific and Innovative Research, CSIR-Human Resource Development Campus, Ghaziabad, India.
- 196 Genomics and Molecular Medicine Unit, CSIR-Institute of Genomics and Integrative Biology, New Delhi, India.
- 197 Department of Preventive Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 198 Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
- 199 Ophthalmology and Visual Sciences Academic Clinical Program (Eye ACP), Duke-NUS Medical School, Singapore, Singapore.
- 200 Department of Ophthalmology, Yong Loo Lin School of Medicine, National University of Singapore and National University Health System, Singapore, Singapore.
- 201 Institute of Cardiovascular and Medical Sciences, University of Glasgow, Glasgow, UK.
- 202 Survey Research Center, Institute for Social Research, University of Michigan, Ann Arbor, MI, USA.
- 203 Institute of Genetic Epidemiology, Helmholtz Zentrum Munchen, German Research Center for Environmental Health, Neuherberg, Germany.
- 204 Institute for Medical Biostatistics, Epidemiology and Informatics (IMBEI), University Medical Center, Johannes Gutenberg University, Mainz, Germany.
- 205 Chair of Genetic Epidemiology, Institute of Medical Information Processing, Biometry, and Epidemiology, Faculty of Medicine, Ludwig Maximilians Universität München, Munich, Germany.
- 206 Faculty of Medicine, University of Iceland, Reykjavik, Iceland.
- 207 Faculty of Medicine, Macau University of Science and Technology, Macau, China.
- 208 Department of Medical Genetics and Medical Research, China Medical University Hospital, Taichung, Taiwan.
- 209 Population Health Unit, Finnish Institute for Health and Welfare, Helsinki, Finland, Finnish Institute for Health and Welfare, Helsinki, Finland.
- 210 National School of Public Health, Madrid, Spain.
- 211 Department of Public Health, University of Helsinki, Helsinki, Finland.
- 212 Diabetes Research Group, King Abdulaziz University, Jeddah, Saudi Arabia.
- 213 Unidad de Biología Molecular y Medicina Genómica, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
- 214 Departamento de Medicina Genómica y Toxiología Ambiental, Instituto de Investigaciones Biomédicas, UNAM, Mexico City, Mexico.
- 215 Unidad de Investigacion Medica en Bioquimica, Hospital de Especialidades, Centro Medico Nacional Siglo XXI, Instituto Mexicano del Seguro Social, Mexico City, Mexico.
- 216 Einthoven Laboratory for Experimental Vascular Medicine, Leiden University Medical Center, Leiden, The Netherlands.
- 217 Department of Human Genetics, Leiden University Medical Center, Leiden, The Netherlands.
- 218 Department of Clinical Chemistry, Laboratory of Genetic Metabolic Disease, Amsterdam University Medical Center, Amsterdam, The Netherlands.
- 219 Southern California Eye Institute, CHA Hollywood Presbyterian Hospital, Los Angeles, CA, USA.
- 220 Unidad de Investigación Médica en Epidemiologia Clinica, Hospital de Especialidades, Centro Medico Nacional Siglo XXI, Instituto Mexicano del Seguro Social, Mexico City, Mexico.
- 221 Department of Internal Medicine, Division of Endocrinology, Leiden University Medical Center, Leiden, The Netherlands.
- 222 Department of Public Health, Aarhus University, Aarhus, Denmark.
- 223 Danish Diabetes Academy, Odense, Denmark.
- 224 Diabetology Research Centre, King Edward Memorial Hospital and Research Centre, Pune, India.
- 225 Department of Medical Biochemistry, Kurume University School of Medicine, Kurume, Japan.
- 226 Department of Pediatrics, Osaka University Graduate School of Medicine, Suita, Japan.
- 227 Division of Cancer Control and Population Sciences, UPMC Hillman Cancer Center, University of Pittsburgh, Pittsburgh, PA, USA.
- 228 Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA.
- 229 Department of Pediatrics, Division of Genetic and Genomic Medicine, UCI Irvine School of Medicine, Irvine, CA, USA.
- 230 Department of Anti-Aging Medicine, Ehime University Graduate School of Medicine, Ehime, Japan.
- 231 Division of Pulmonary, Critical Care, and Sleep Medicine, Beth Israel Deaconess Medical Center, Boston, MA, USA.
- 232 Department of Medical Sciences, Uppsala University, Uppsala, Sweden.
- 233 Institute of Molecular Medicine, The University of Texas Health Science Center at Houston School of Public Health, Houston, TX, USA.
- 234 Human Genetics Center, University of Texas Health Science Center at Houston, Houston, TX, US.
- 235 Department of Medicine, Stanford University School of Medicine, Stanford, CA, USA.
- 236 Department of Medical Sciences, Molecular Epidemiology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden.
- 237 Department of Epidemiology, University of Washington, Seattle, WA, USA.
- 238 Department of Health Systems and Population Health, University of Washington, Seattle, WA, USA.
- 239 Beijing Institute of Ophthalmology, Ophthalmology and Visual Sciences Key Laboratory, Beijing Tongren Hospital, Capital Medical University, Beijing, China.
- 240 Department of Medicine, Division of Cardiology, Duke University School of Medicine, Durham, NC, USA.
- 241 Kurume University School of Medicine, Kurume, Japan.
- 242 Department of Medicine, McGill University, Montreal, QC, Canada.
- 243 Department of Human Genetics, McGill University, Montreal, QC, Canada.
- 244 Department of Metabolism, Digestion and Reproduction, Imperial College London, London, UK.
- 245 Department of Physiology and Biophysics, University of Mississippi Medical Center, Jackson, MS, USA.
- 246 Division of Endocrinology and Metabolism, Department of Medicine, Taichung Veterans General Hospital, Taichung, Taiwan.
- 247 Center for Genetic Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
- 248 Department of Anthropology, Northwestern University, Evanston, IL, USA.
- 249 Department of Endocrinology, Helsinki University Hospital, Helsinki, Finland.
- 250 Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland.
- 251 Folkhalsan Research Center, Helsinki, Finland.
- 252 Lund University Diabetes Centre, Malmö, Sweden.
- 253 Systems Genomics Laboratory, School of Biotechnology, Jawaharlal Nehru University, New Delhi, India.
- 254 Department of Pathology and Molecular Medicine, McMaster University, Hamilton, ON, Canada.
- 255 Deceased.
- 256 Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, South Korea.
- 257 Netherlands Heart Institute, Utrecht, The Netherlands.
- 258 Center for Public Health Genomics, University of Virginia School of Medicine, Charlottesville, VA, USA.
- 259 Research Unit of Molecular Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Munich, Germany.
- 260 Duke-NUS Medical School, Singapore, Singapore.
- 261 Department of Epidemiology, Biostatistics and Occupational Health, McGill University, Montreal, QC, Canada.
- 262 Singapore Institute for Clinical Sciences, Agency for Science Technology and Research (A*STAR), Singapore, Singapore.
- 263 Healthy Longevity Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
- 264 Center for Diabetes Research, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 265 Department of Biochemistry, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 266 Pat Macpherson Centre for Pharmacogenetics and Pharmacogenomics, University of Dundee, Dundee, UK.
- 267 Imperial College Healthcare NHS Trust, Imperial College London, London, UK.
- 268 MRC-PHE Centre for Environment and Health, Imperial College London, London, UK.
- 269 National Heart and Lung Institute, Imperial College London, London, UK.
- 270 Department of Medicine, Brown University Alpert School of Medicine, Providence, RI, USA.
- 271 Department of Medicine, Columbia University Irving Medical Center, New York, NY, USA.
- 272 Department of Cardiology, Columbia University Irving Medical Center, New York, NY, USA.
- 273 Center for Non-Communicable Diseases, Karachi, Pakistan.
- 274 Computational Medicine, Berlin Institute of Health at Charité Universitätsmedizin, Berlin, Germany.
- 275 Precision Healthcare University Research Institute, Queen Mary University of London, London, UK.
- 276 Department of Preventive Medicine, Keck School of Medicine of USC, Los Angeles, CA, USA.
- 277 The Mindich Child Health and Development Institute, Ichan School of Medicine at Mount Sinai, New York, NY, USA.
- 278 Division of Translational Medicine and Therapeutics, Department of Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 279 Institute for Translational Medicine and Therapeutics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 280 Department of Pediatrics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 281 Center for Precision Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 282 Institute for Biomedical Informatics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 283 Department of Psychiatry, University of Michigan, Ann Arbor, MI, USA.
- 284 All of Us Research Program, National Institutes of Health, Bethesda, MD, USA.
- 285 Toranomon Hospital, Tokyo, Japan.
- 286 Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore.
- 287 Vanderbilt Genetics Institute, Division of Genetic Medicine, Vanderbilt University Medical Center, Nashville, TN, USA.
- 288 VA Palo Alto Health Care System, Palo Alto, CA, USA.
- 289 Stanford Cardiovascular Institute, Stanford University School of Medicine, Stanford, CA, USA.
- 290 Department of Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 291 Oxford Centre for Diabetes, Endocrinology and Metabolism, Radcliffe Department of Medicine, University of Oxford, Oxford, UK.
- 292 Oxford NIHR Biomedical Research Centre, Churchill Hosptial, Oxford University Hospitals NHS Foundation Trust, Oxford, UK.
- 293 Present address: Genentech, South San Francisco, CA, USA.
- 294 Department of Biostatistics and Epidemiology, University of Massachusetts Amherst, Amherst, MA, USA.
- 295 Division of Epidemiology, Department of Biostatistics, Epidemiology and Informatics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
- 296 TUM School of Medicine, Technical University of Munich and Klinikum Rechts der Isar, Munich, Germany.
- PMID: 37034649
- PMCID: PMC10081410
- DOI: 10.1101/2023.03.31.23287839
Abstract
Type 2 diabetes (T2D) is a heterogeneous disease that develops through diverse pathophysiological processes. To characterise the genetic contribution to these processes across ancestry groups, we aggregate genome-wide association study (GWAS) data from 2,535,601 individuals (39.7% non-European ancestry), including 428,452 T2D cases. We identify 1,289 independent association signals at genome-wide significance (P<5×10-8) that map to 611 loci, of which 145 loci are previously unreported. We define eight non-overlapping clusters of T2D signals characterised by distinct profiles of cardiometabolic trait associations. These clusters are differentially enriched for cell-type specific regions of open chromatin, including pancreatic islets, adipocytes, endothelial, and enteroendocrine cells. We build cluster-specific partitioned genetic risk scores (GRS) in an additional 137,559 individuals of diverse ancestry, including 10,159 T2D cases, and test their association with T2D-related vascular outcomes. Cluster-specific partitioned GRS are more strongly associated with coronary artery disease and end-stage diabetic nephropathy than an overall T2D GRS across ancestry groups, highlighting the importance of obesity-related processes in the development of vascular outcomes. Our findings demonstrate the value of integrating multi-ancestry GWAS with single-cell epigenomics to disentangle the aetiological heterogeneity driving the development and progression of T2D, which may offer a route to optimise global access to genetically-informed diabetes care.
Conflict of interest statement
R.A.S. is now an employee of GlaxoSmithKline. G.T. is an employee of deCODE genetics/Amgen Inc. A.S.B. reports institutional grants from AstraZeneca, Bayer, Biogen, BioMarin, Bioverativ, Novartis, Regeneron and Sanofi. J.Danesh serves on scientific advisory boards for AstraZeneca, Novartis, and UK Biobank, and has received multiple grants from academic, charitable and industry sources outside of the submitted work. L.S.E. is now an employee of Bristol Myers Squibb. J.S.F. has consulted for Shionogi Inc. T.M.F. has consulted for Sanofi, Boehringer Ingelheim, and received funding from GlaxoSmithKline. H.C.G. holds the McMaster-Sanofi Population Health Institute Chair in Diabetes Research and Care; reports research grants from Eli Lilly, AstraZeneca, Merck, Novo Nordisk and Sanofi; honoraria for speaking from AstraZeneca, Boehringer Ingelheim, Eli Lilly, Novo Nordisk, DKSH, Zuellig, Roche, and Sanofi; and consulting fees from Abbott, AstraZeneca, Boehringer Ingelheim, Eli Lilly, Merck, Novo Nordisk, Pfizer, Sanofi, Kowa and Hanmi.lth Institute Chair in Diabetes Research and Care; reports research grants from Eli Lilly, AstraZeneca, Merck, Novo Nordisk and Sanofi; honoraria for speaking from AstraZeneca, Boehringer Ingelheim, Eli Lilly, Novo Nordisk, and Sanofi; and consulting fees from Abbott, AstraZeneca, Boehringer Ingelheim, Eli Lilly, Merck, Novo Nordisk, Janssen, Sanofi, and Kowa. M.Ingelsson is a paid consultant to BioArctic AB. R.L.-G. is a part-time consultant of Metabolon Inc. A.E.L. is now an employee of Regeneron Genetics Center LLC and holds shares in Regeneron Pharmaceuticals. M.A.N. currently serves on the scientific advisory board for Clover Therapeutics and is an advisor to Neuron23 Inc. S.R.P. has received grant funding from Bayer Pharmaceuticals, Philips Respironics and Respicardia. N.Sattar has consulted for or been on speakers bureau for Abbott, Amgen, Astrazeneca, Boehringer Ingelheim, Eli Lilly, Hanmi, Novartis, Novo Nordisk, Sanofi and Pfizer and has received grant funding from Astrazeneca, Boehringer Ingelheim, Novartis and Roche Diagnostics. V.S. is now an employee of deCODE genetics/Amgen Inc. A.M.S. receives funding from Seven Bridges Genomics to develop tools for the NHLBI BioData Catalyst consortium. U.T. is an employee of deCODE genetics/Amgen Inc. E.Ingelsson is now an employee of GlaxoSmithKline. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. R.C.W.M. reports research funding from AstraZeneca, Bayer, Novo Nordisk, Pfizer, Tricida Inc. and Sanofi, and has consulted for or received speakers fees from AstraZeneca, Bayer, Boehringer Ingelheim, all of which have been donated to the Chinese University of Hong Kong to support diabetes research. D.O.M.-K. is a part-time clinical research consultant for Metabolon Inc. S.Liu reports consulting payments and honoraria or promises of the same for scientific presentations or reviews at numerous venues, including but not limited to Barilla, by-Health Inc, Ausa Pharmed Co.LTD, Fred Hutchinson Cancer Center, Harvard University, University of Buffalo, Guangdong General Hospital and Academy of Medical Sciences, Consulting member for Novo Nordisk, Inc; member of the Data Safety and Monitoring Board for a trial of pulmonary hypertension in diabetes patients at Massachusetts General Hospital; receives royalties from UpToDate; receives an honorarium from the American Society for Nutrition for his duties as Associate Editor. K.Stefansson is an employee of deCODE genetics/Amgen Inc. M.I.M. has served on advisory panels for Pfizer, NovoNordisk and Zoe Global, has received honoraria from Merck, Pfizer, Novo Nordisk and Eli Lilly, and research funding from Abbvie, Astra Zeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, NovoNordisk, Pfizer, Roche, Sanofi Aventis, Servier, and Takeda; and is now an employee of Genentech and a holder of Roche stock. J.B.M. is an Academic Associate for Quest Diagnostics R&D. A.Mahajan is an employee of Genentech, and a holder of Roche stock.
Figures
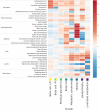
Figure 1.. Heatmap of associations of 37…
Figure 1.. Heatmap of associations of 37 cardiometabolic phenotypes with eight mechanistic clusters of index…
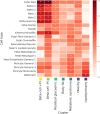
Figure 2.. Heatmap of cluster-specific enrichments of…
Figure 2.. Heatmap of cluster-specific enrichments of T2D associations for cell type-specific regions of open…

Figure 3.. Associations of the overall GRS…
Figure 3.. Associations of the overall GRS and cluster-specific partitioned GRS with five T2D-related vascular…
References
-
- International Diabetes Federation. IDF Diabetes Atlas. 10th edn., Brussels, Belgium: (2021). https://diabetesatlas.org/.
-
- Almgren P. et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia 54, 2811–2819 (2011). - PubMed
Publication types
Grants and funding
- UM1 DK126194/DK/NIDDK NIH HHS/United States
- R00 AG066849/AG/NIA NIH HHS/United States
- R03 DK131249/DK/NIDDK NIH HHS/United States
- R01 HD030880/HD/NICHD NIH HHS/United States
- K23 DK114551/DK/NIDDK NIH HHS/United States
- R01 AG065357/AG/NIA NIH HHS/United States
- U01 HG011723/HG/NHGRI NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- MC_UU_00006/1/MRC_/Medical Research Council/United Kingdom
- 212945/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- R01 HL153805/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- U01 DK105535/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
