This is a preprint.
Deep repertoire mining uncovers ultra-broad coronavirus neutralizing antibodies targeting multiple spike epitopes
- PMID: 37034676
- PMCID: PMC10081229
- DOI: 10.1101/2023.03.28.534602
Deep repertoire mining uncovers ultra-broad coronavirus neutralizing antibodies targeting multiple spike epitopes
Update in
-
Deep repertoire mining uncovers ultra-broad coronavirus neutralizing antibodies targeting multiple spike epitopes.Cell Rep. 2024 Jun 25;43(6):114307. doi: 10.1016/j.celrep.2024.114307. Epub 2024 Jun 5. Cell Rep. 2024. PMID: 38848216 Free PMC article.
Abstract
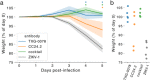
Development of vaccines and therapeutics that are broadly effective against known and emergent coronaviruses is an urgent priority. Current strategies for developing pan-coronavirus countermeasures have largely focused on the receptor binding domain (RBD) and S2 regions of the coronavirus Spike protein; it has been unclear whether the N-terminal domain (NTD) is a viable target for universal vaccines and broadly neutralizing antibodies (Abs). Additionally, many RBD-targeting Abs have proven susceptible to viral escape. We screened the circulating B cell repertoires of COVID-19 survivors and vaccinees using multiplexed panels of uniquely barcoded antigens in a high-throughput single cell workflow to isolate over 9,000 SARS-CoV-2-specific monoclonal Abs (mAbs), providing an expansive view of the SARS-CoV-2-specific Ab repertoire. We observed many instances of clonal coalescence between individuals, suggesting that Ab responses frequently converge independently on similar genetic solutions. Among the recovered antibodies was TXG-0078, a public neutralizing mAb that binds the NTD supersite region of the coronavirus Spike protein and recognizes a diverse collection of alpha- and beta-coronaviruses. TXG-0078 achieves its exceptional binding breadth while utilizing the same VH1-24 variable gene signature and heavy chain-dominant binding pattern seen in other NTD supersite-specific neutralizing Abs with much narrower specificity. We also report the discovery of CC24.2, a pan-sarbecovirus neutralizing mAb that targets a novel RBD epitope and shows similar neutralization potency against all tested SARS-CoV-2 variants, including BQ.1.1 and XBB.1.5. A cocktail of TXG-0078 and CC24.2 provides protection against in vivo challenge with SARS-CoV-2, suggesting potential future use in variant-resistant therapeutic Ab cocktails and as templates for pan-coronavirus vaccine design.
Conflict of interest statement
Competing interests D.B.J., B.A.A., M.J.T.S., and W.J.M. were employees and shareholders of 10x Genomics, Inc. at the time this work was performed. D.B.J., B.A.A., M.J.T.S., and W.J.M. are inventors on patents in relation to algorithms, therapeutic candidates, and other components of this work.
Figures


References
-
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348: 1967–1976. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
