This is a preprint.
RecA-NT homology motif in ImuB is essential for mycobacterial ImuA'-ImuB protein interaction and mutasome function
- PMID: 37034714
- PMCID: PMC10081233
- DOI: 10.1101/2023.03.28.534377
RecA-NT homology motif in ImuB is essential for mycobacterial ImuA'-ImuB protein interaction and mutasome function
Update in
-
The RecA-NT homology motif in ImuB mediates the interaction with ImuA', which is essential for DNA damage-induced mutagenesis.J Biol Chem. 2025 Feb;301(2):108108. doi: 10.1016/j.jbc.2024.108108. Epub 2024 Dec 18. J Biol Chem. 2025. PMID: 39706264 Free PMC article.
Abstract
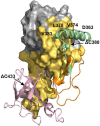
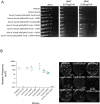
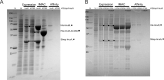
The mycobacterial mutasome - minimally comprising ImuA', ImuB, and DnaE2 proteins - has been implicated in DNA damage-induced mutagenesis in Mycobacterium tuberculosis. ImuB, predicted to enable mutasome function via its interaction with the β clamp, is a catalytically inactive member of the Y-family of DNA polymerases. Like other members of the Y family, ImuB features a recently identified amino acid motif with homology to the RecA-N-terminus (RecA-NT). In RecA, the motif mediates oligomerization of RecA monomers into RecA filaments. Given the role of ImuB in the mycobacterial mutasome, we hypothesized that the ImuB RecA-NT motif might mediate its interaction with ImuA', a RecA homolog of unknown function. To investigate this possibility, we constructed a panel of imuB alleles in which RecA-NT was removed, or mutated. Results from microbiological and biochemical assays indicate that RecA-NT is critical for the interaction of ImuB with ImuA'. A region downstream of RecA-NT (ImuB-C) also appears to stabilize the ImuB-ImuA' interaction, but its removal does not prevent complex formation. In contrast, replacing two key hydrophobic residues of RecA-NT, L378 and V383, is sufficient to disrupt ImuA'-ImuB interaction. To our knowledge, this constitutes the first experimental evidence showing the role of the RecA-NT motif in mediating the interaction between a Y-family member and a RecA homolog.
Conflict of interest statement
CONFLICT OF INTEREST The authors declare that they have no competing interests.
Figures

Similar articles
-
The RecA-NT homology motif in ImuB mediates the interaction with ImuA', which is essential for DNA damage-induced mutagenesis.J Biol Chem. 2025 Feb;301(2):108108. doi: 10.1016/j.jbc.2024.108108. Epub 2024 Dec 18. J Biol Chem. 2025. PMID: 39706264 Free PMC article.
-
Essential roles for imuA'- and imuB-encoded accessory factors in DnaE2-dependent mutagenesis in Mycobacterium tuberculosis.Proc Natl Acad Sci U S A. 2010 Jul 20;107(29):13093-8. doi: 10.1073/pnas.1002614107. Epub 2010 Jul 6. Proc Natl Acad Sci U S A. 2010. PMID: 20615954 Free PMC article.
-
Investigating the composition and recruitment of the mycobacterial ImuA'-ImuB-DnaE2 mutasome.Elife. 2023 Aug 2;12:e75628. doi: 10.7554/eLife.75628. Elife. 2023. PMID: 37530405 Free PMC article.
-
A simple recipe for the non-expert bioinformaticist for building experimentally-testable hypotheses for proteins with no known homologs.J Struct Funct Genomics. 2012 Dec;13(4):185-200. doi: 10.1007/s10969-012-9141-7. Epub 2012 Sep 7. J Struct Funct Genomics. 2012. PMID: 22956349 Review.
-
Coexistence of SOS-Dependent and SOS-Independent Regulation of DNA Repair Genes in Radiation-Resistant Deinococcus Bacteria.Cells. 2021 Apr 16;10(4):924. doi: 10.3390/cells10040924. Cells. 2021. PMID: 33923690 Free PMC article. Review.
References
-
- Boshoff H.I.M.M., Reed M.B., Barry C.E. and Mizrahi V. (2003) DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell, 113(2),183–193. - PubMed
-
- Gessner S., Martin Z., Reiche M.A., Santos J.A., Dhar N., Dinkele R., De Wet T., Ramudzuli A., Anoosheh S., Lang D.M., et al. (2021) The mycobacterial mutasome: composition and recruitment in live cells. bioRxiv doi: 10.1101/2021.11.16.468908, 17 November 2021, pre-print:not peer-reviewed. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
