This is a preprint.
APOBEC3 degradation is the primary function of HIV-1 Vif for virus replication in the myeloid cell line THP-1
- PMID: 37034786
- PMCID: PMC10081227
- DOI: 10.1101/2023.03.28.534666
APOBEC3 degradation is the primary function of HIV-1 Vif for virus replication in the myeloid cell line THP-1
Update in
-
APOBEC3 degradation is the primary function of HIV-1 Vif determining virion infectivity in the myeloid cell line THP-1.mBio. 2023 Aug 31;14(4):e0078223. doi: 10.1128/mbio.00782-23. Epub 2023 Aug 9. mBio. 2023. PMID: 37555667 Free PMC article.
Abstract
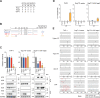
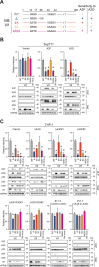
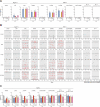
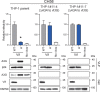
HIV-1 must overcome multiple innate antiviral mechanisms to replicate in CD4 + T lymphocytes and macrophages. Previous studies have demonstrated that the APOBEC3 (A3) family of proteins (at least A3D, A3F, A3G, and stable A3H haplotypes) contribute to HIV-1 restriction in CD4 + T lymphocytes. Virus-encoded virion infectivity factor (Vif) counteracts this antiviral activity by degrading A3 enzymes allowing HIV-1 replication in infected cells. In addition to A3 proteins, Vif also targets other cellular proteins in CD4 + T lymphocytes, including PPP2R5 proteins. However, whether Vif primarily degrades only A3 proteins or has additional essential targets during viral replication is currently unknown. Herein, we describe the development and characterization of A3F -, A3F/A3G -, and A3A -to- A3G -null THP-1 cells. In comparison to Vif-proficient HIV-1, Vif-deficient viruses have substantially reduced infectivity in parental and A3F -null THP-1 cells, and a more modest decrease in infectivity in A3F/A3G -null cells. Remarkably, disruption of A3Aâ€"A3G protein expression completely restores the infectivity of Vif-deficient viruses in THP-1 cells. These results indicate that the primary function of Vif during HIV-1 replication in THP-1 cells is the targeting and degradation of A3 enzymes.
Importance: HIV-1 Vif neutralizes the HIV-1 restriction activity of A3 proteins. However, it is currently unclear whether Vif has additional essential cellular targets. To address this question, we disrupted A3A to A3G genes in the THP-1 myeloid cell line using CRISPR and compared the infectivity of wildtype HIV-1 and Vif mutants with the selective A3 neutralization activities. Our results demonstrate that the infectivity of Vif-deficient HIV-1 and the other Vif mutants is fully restored by ablating the expression of cellular A3A to A3G proteins. These results indicate that A3 proteins are the only essential target of Vif that is required for HIV-1 replication in THP-1 cells.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
APOBEC3 degradation is the primary function of HIV-1 Vif determining virion infectivity in the myeloid cell line THP-1.mBio. 2023 Aug 31;14(4):e0078223. doi: 10.1128/mbio.00782-23. Epub 2023 Aug 9. mBio. 2023. PMID: 37555667 Free PMC article.
-
HIV-1 and HIV-2 Vif interact with human APOBEC3 proteins using completely different determinants.J Virol. 2014 Sep 1;88(17):9893-908. doi: 10.1128/JVI.01318-14. Epub 2014 Jun 18. J Virol. 2014. PMID: 24942576 Free PMC article.
-
APOBEC3G and APOBEC3F Act in Concert To Extinguish HIV-1 Replication.J Virol. 2016 Apr 14;90(9):4681-4695. doi: 10.1128/JVI.03275-15. Print 2016 May. J Virol. 2016. PMID: 26912618 Free PMC article.
-
Dual Functionality of HIV-1 Vif in APOBEC3 Counteraction and Cell Cycle Arrest.Front Microbiol. 2021 Jan 12;11:622012. doi: 10.3389/fmicb.2020.622012. eCollection 2020. Front Microbiol. 2021. PMID: 33510734 Free PMC article. Review.
-
Structural Insights into APOBEC3-Mediated Lentiviral Restriction.Viruses. 2020 May 27;12(6):587. doi: 10.3390/v12060587. Viruses. 2020. PMID: 32471198 Free PMC article. Review.
References
-
- Koito A, Ikeda T. 2012. Apolipoprotein B mRNA-editing, catalytic polypeptide cytidine deaminases and retroviral restriction. Wiley Interdiscip Rev RNA 3:529–41. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
