Addition of dNTPs can improve the detection sensitivity of catalytic hairpin assembly
- PMID: 37034995
- PMCID: PMC10074145
- DOI: 10.1016/j.isci.2023.106390
Addition of dNTPs can improve the detection sensitivity of catalytic hairpin assembly
Abstract
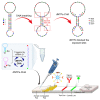
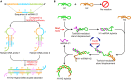
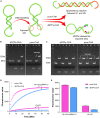
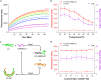
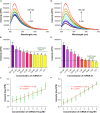
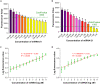
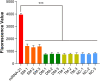
Ever since the catalytic hairpin assembly (CHA) circuit has been highlighted as a powerful nucleic acid detection tool, nucleic acid detection methods based on CHA have been widely studied. However, the detection sensitivity of CHA-based methods is insufficient. The relatively high background signals resulting from the spontaneous reaction between the two hairpin probes is one of the major reasons for limiting the sensitivity of CHA. In this study, we established that the addition of deoxynucleotide triphosphates (dNTPs) to the reaction system can significantly reduce the background leakage of CHA. The dNTPs-CHA, coupled with a fluorescence lateral flow assay strip, is used for the rapid and highly sensitive detection of miRNA. It is capable of reliably detecting miRNA in serum samples down to a limit of 100 aM, which is an improvement in the lower detection limit by nearly five orders of magnitude compared to that of the pure CHA.
Keywords: Biological sciences; Diagnostics; Molecular biology.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources

