Identification of a centrosome-related prognostic signature for breast cancer
- PMID: 37035151
- PMCID: PMC10073657
- DOI: 10.3389/fonc.2023.1138049
Identification of a centrosome-related prognostic signature for breast cancer
Abstract
Background: As the major microtubule organizing center in animal cells, the centrosome is implicated with human breast tumor in multiple ways, such as promotion of tumor cell immune evasion. Here, we aimed to detect the expression of centrosome-related genes (CRGs) in normal and malignant breast tissues, and construct a novel centrosome-related prognostic model to discover new biomarkers and screen drugs for breast cancer.
Methods: We collected CRGs from the public databases and literature. The differentially expressed CRGs between normal and malignant breast tissues were identified by the DESeq2. Univariate Cox and LASSO regression analyses were conducted to screen candidate prognostic CRGs and develop a centrosome-related signature (CRS) to score breast cancer patients. We further manipulated and visualized data from TCGA, GEO, IMvigor210, TCIA and TIMER to explore the correlation between CRS and patient outcomes, clinical manifestations, mutational landscapes, tumor immune microenvironments, and responses to diverse therapies. Single cell analyses were performed to investigate the difference of immune cell landscape between high- and low-risk group patients. In addition, we constructed a nomogram to guide clinicians in precise treatment.
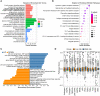
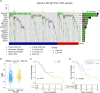
Results: A total of 726 CRGs were collected from the public databases and literature. PSME2, MAPK10, EIF4EBP1 were screened as the prognostic genes in breast cancer. Next, we constructed a centrosome-related prognostic signature and validated its efficacy based on the genes for predicting the survival of breast cancer patients. The high-risk group patients had poor prognoses, the area under the ROC curve for 1-, 3-, and 5-year overall survival (OS) was 0.77, 0.67, and 0.65, respectively. The predictive capacity of CRS was validated by other datasets from GEO dataset. In addition, high-risk group patients exhibited elevated level of mutational landscapes and decreased level of immune infiltration, especially T and B lymphocytes. In terms of treatment responses, patients in the high-risk group were found to be resistant to immunotherapy but sensitive to chemotherapy. Moreover, we screened a series of candidate anticancer drugs with high sensitivity in the high-risk group.
Conclusion: Our work exploited a centrosome-related prognostic signature and developed a predictive nomogram capable of accurately predicting breast cancer OS. The above discoveries provide deeper insights into the vital roles of the centrosome and contribute to the development of personalized treatment for breast cancer.
Keywords: CRSS; breast cancer; centrosome; immune; machine algorithm; prognostic model.
Copyright © 2023 Fang, Gao, Yu, Sun and Yao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Burstein HJ, Curigliano G, Thürlimann B, Weber WP, Poortmans P, Regan MM, et al. Customizing local and systemic therapies for women with early breast cancer: the St. Gallen international consensus guidelines for treatment of early breast cancer 2021. Ann Oncol (2021) 32(10):1216–35. doi: 10.1016/j.annonc.2021.06.023 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

