YB1 participated in regulating mitochondrial activity through RNA replacement
- PMID: 37035211
- PMCID: PMC10076880
- DOI: 10.3389/fonc.2023.1145379
YB1 participated in regulating mitochondrial activity through RNA replacement
Abstract
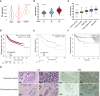
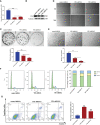
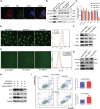
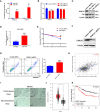
As a relic of ancient bacterial endosymbionts, mitochondria play a central role in cell metabolism, apoptosis, autophagy, and other processes. However, the function of mitochondria-derived nucleic acids in cellular signal transduction has not been fully elucidated. Here, our work has found that Y-box binding protein 1 (YB1) maintained cellular autophagy at a moderate level to inhibit mitochondrial oxidative phosphorylation. In addition, mitochondrial RNA was leaked into cytosol under starvation, accompanied by YB1 mitochondrial relocation, resulting in YB1-bound RNA replacement. The mRNAs encoded by oxidative phosphorylation (OXPHOS)-associated genes and oncogene HMGA1 (high-mobility group AT-hook 1) were competitively replaced by mitochondria-derived tRNAs. The increase of free OXPHOS mRNAs released from the YB1 complex enhanced mitochondrial activity through facilitating translation, but the stability of HMGA1 mRNA was impaired without the protection of YB1, both contributing to breast cancer cell apoptosis and reactive oxygen species production. Our finding not only provided a new potential target for breast cancer therapy but also shed new light on understanding the global landscape of cellular interactions between RNA-binding proteins and different RNA species.
Keywords: HMGA1; YB1; apoptosis; autophagy; mitochondria.
Copyright © 2023 Gong and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
BRD7 suppresses invasion and metastasis in breast cancer by negatively regulating YB1-induced epithelial-mesenchymal transition.J Exp Clin Cancer Res. 2020 Feb 7;39(1):30. doi: 10.1186/s13046-019-1493-4. J Exp Clin Cancer Res. 2020. PMID: 32028981 Free PMC article.
-
YB1 protects cardiac myocytes against H2O2‑induced injury via suppression of PIAS3 mRNA and phosphorylation of STAT3.Mol Med Rep. 2019 Jun;19(6):4579-4588. doi: 10.3892/mmr.2019.10119. Epub 2019 Apr 3. Mol Med Rep. 2019. PMID: 30942400 Free PMC article.
-
Ribonucleoprotein Y-box-binding protein-1 regulates mitochondrial oxidative phosphorylation (OXPHOS) protein expression after serum stimulation through binding to OXPHOS mRNA.Biochem J. 2012 Apr 15;443(2):573-84. doi: 10.1042/BJ20111728. Biochem J. 2012. PMID: 22280412
-
YB1 and its role in osteosarcoma: a review.Front Oncol. 2024 Oct 21;14:1452661. doi: 10.3389/fonc.2024.1452661. eCollection 2024. Front Oncol. 2024. PMID: 39497723 Free PMC article. Review.
-
Reprogramming Oxidative Phosphorylation in Cancer: A Role for RNA-Binding Proteins.Antioxid Redox Signal. 2020 Nov 1;33(13):927-945. doi: 10.1089/ars.2019.7988. Epub 2020 Jan 30. Antioxid Redox Signal. 2020. PMID: 31910046 Review.
Cited by
-
Knockdown of Y-box binding protein 1 induces autophagy in early porcine embryos.Front Cell Dev Biol. 2023 Oct 30;11:1238546. doi: 10.3389/fcell.2023.1238546. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37965572 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

