Dual inhibition of EZH2 and G9A/GLP histone methyltransferases by HKMTI-1-005 promotes differentiation of acute myeloid leukemia cells
- PMID: 37035245
- PMCID: PMC10076884
- DOI: 10.3389/fcell.2023.1076458
Dual inhibition of EZH2 and G9A/GLP histone methyltransferases by HKMTI-1-005 promotes differentiation of acute myeloid leukemia cells
Abstract
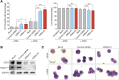
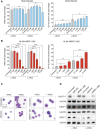
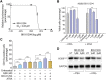
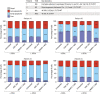
All-trans-retinoic acid (ATRA)-based differentiation therapy of acute promyelocytic leukemia (APL) represents one of the most clinically effective examples of precision medicine and the first example of targeted oncoprotein degradation. The success of ATRA in APL, however, remains to be translated to non-APL acute myeloid leukemia (AML). We previously showed that aberrant histone modifications, including histone H3 lysine 4 (H3K4) and lysine 27 (H3K27) methylation, were associated with this lack of response and that epigenetic therapy with small molecule inhibitors of the H3K4 demethylase LSD1/KDM1A could reprogram AML cells to respond to ATRA. Serving as the enzymatic component of Polycomb Repressive Complex 2, EZH2/KMT6A methyltransferase plays a critical role in normal hematopoiesis by affecting the balance between self-renewal and differentiation. The canonical function of EZH2 is methylation of H3K27, although important non-canonical roles have recently been described. EZH2 mutation or deregulated expression has been conclusively demonstrated in the pathogenesis of AML and response to treatment, thus making it an attractive therapeutic target. In this study, we therefore investigated whether inhibition of EZH2 might also improve the response of non-APL AML cells to ATRA-based therapy. We focused on GSK-343, a pyridone-containing S-adenosyl-L-methionine cofactor-competitive EZH2 inhibitor that is representative of its class, and HKMTI-1-005, a substrate-competitive dual inhibitor targeting EZH2 and the closely related G9A/GLP H3K9 methyltransferases. We found that treatment with HKMTI-1-005 phenocopied EZH2 knockdown and was more effective in inducing differentiation than GSK-343, despite the efficacy of GSK-343 in terms of abolishing H3K27 trimethylation. Furthermore, transcriptomic analysis revealed that in contrast to treatment with GSK-343, HKMTI-1-005 upregulated the expression of differentiation pathway genes with and without ATRA, while downregulating genes associated with a hematopoietic stem cell phenotype. These results pointed to a non-canonical role for EZH2, which was supported by the finding that EZH2 associates with the master regulator of myeloid differentiation, RARα, in an ATRA-dependent manner that was enhanced by HKMTI-1-005, possibly playing a role in co-regulator complex exchange during transcriptional activation. In summary, our results strongly suggest that addition of HKMTI-1-005 to ATRA is a new therapeutic approach against AML that warrants further investigation.
Keywords: EZH2/KMT6A; G9A/EHMT2; GLP/EHMT1; HKMTI-1-005; RARα; acute myeloid leukemia (AML); all-trans retinoic acid (ATRA); differentiation.
Copyright © 2023 Sbirkov, Schenk, Kwok, Stengel, Brown, Brown, Chesler, Zelent, Fuchter and Petrie.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia.Nat Med. 2012 Mar 11;18(4):605-11. doi: 10.1038/nm.2661. Nat Med. 2012. PMID: 22406747 Free PMC article.
-
Knockdown of SALL4 Protein Enhances All-trans Retinoic Acid-induced Cellular Differentiation in Acute Myeloid Leukemia Cells.J Biol Chem. 2015 Apr 24;290(17):10599-609. doi: 10.1074/jbc.M114.634790. Epub 2015 Mar 3. J Biol Chem. 2015. PMID: 25737450 Free PMC article.
-
Sensitivity of MLL-rearranged AML cells to all-trans retinoic acid is associated with the level of H3K4me2 in the RARα promoter region.Blood Cancer J. 2014 Apr 25;4(4):e205. doi: 10.1038/bcj.2014.25. Blood Cancer J. 2014. PMID: 24769646 Free PMC article.
-
Reprogramming acute myeloid leukemia into sensitivity for retinoic-acid-driven differentiation.Exp Hematol. 2017 Aug;52:12-23. doi: 10.1016/j.exphem.2017.04.007. Epub 2017 Apr 27. Exp Hematol. 2017. PMID: 28456748 Review.
-
All-trans retinoic acid in non-promyelocytic acute myeloid leukemia: driver lesion dependent effects on leukemic stem cells.Cell Cycle. 2020 Oct;19(20):2573-2588. doi: 10.1080/15384101.2020.1810402. Epub 2020 Sep 8. Cell Cycle. 2020. PMID: 32900260 Free PMC article. Review.
Cited by
-
How CBX proteins regulate normal and leukemic blood cells.FEBS Lett. 2024 Nov;598(22):2788-2806. doi: 10.1002/1873-3468.14839. Epub 2024 Mar 1. FEBS Lett. 2024. PMID: 38426219 Free PMC article. Review.
-
The epigenetic role of EZH2 in acute myeloid leukemia.PeerJ. 2024 Dec 6;12:e18656. doi: 10.7717/peerj.18656. eCollection 2024. PeerJ. 2024. PMID: 39655332 Free PMC article. Review.
-
Pharmacological targeting of CBX7 alters the epigenetic landscape and induces differentiation of leukemic cells.Blood Neoplasia. 2024 Oct 24;1(4):100052. doi: 10.1016/j.bneo.2024.100052. eCollection 2024 Dec. Blood Neoplasia. 2024. PMID: 40552137 Free PMC article.
-
Epigenetic therapies targeting histone lysine methylation: complex mechanisms and clinical challenges.J Clin Invest. 2024 Oct 15;134(20):e183391. doi: 10.1172/JCI183391. J Clin Invest. 2024. PMID: 39403928 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

