Characterization of the Sterol 24-C-Methyltransferase Genes Reveals a Network of Alternative Sterol Biosynthetic Pathways in Mucor lusitanicus
- PMID: 37036336
- PMCID: PMC10269636
- DOI: 10.1128/spectrum.00315-23
Characterization of the Sterol 24-C-Methyltransferase Genes Reveals a Network of Alternative Sterol Biosynthetic Pathways in Mucor lusitanicus
Abstract
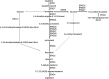
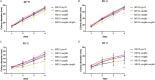
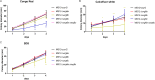
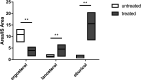
Certain members of the order Mucorales can cause a life-threatening, often-fatal systemic infection called mucormycosis. Mucormycosis has a high mortality rate, which can reach 96 to 100% depending on the underlying condition of the patient. Mucorales species are intrinsically resistant to most antifungal agents, such as most of the azoles, which makes mucormycosis treatment challenging. The main target of azoles is the lanosterol 14α-demethylase (Erg11), which is responsible for an essential step in the biosynthesis of ergosterol, the main sterol component of the fungal membrane. Mutations in the erg11 gene can be associated with azole resistance; however, resistance can also be mediated by loss of function or mutation of other ergosterol biosynthetic enzymes, such as the sterol 24-C-methyltransferase (Erg6). The genome of Mucor lusitanicus encodes three putative erg6 genes (i.e., erg6a, erg6b, and erg6c). In this study, the role of erg6 genes in azole resistance of Mucor was analyzed by generating and analyzing knockout mutants constructed using the CRISPR-Cas9 technique. Susceptibility testing of the mutants suggested that one of the three genes, erg6b, plays a crucial role in the azole resistance of Mucor. The sterol composition of erg6b knockout mutants was significantly altered compared to that of the original strain, and it revealed the presence of at least four alternative sterol biosynthesis pathways leading to formation of ergosterol and other alternative, nontoxic sterol products. Dynamic operation of these pathways and the switching of biosynthesis from one to the other in response to azole treatment could significantly contribute to avoiding the effects of azoles by these fungi. IMPORTANCE The fungal membrane contains ergosterol instead of cholesterol, which offers a specific point of attack for the defense against pathogenic fungi. Indeed, most antifungal agents target ergosterol or its biosynthesis. Mucormycoses-causing fungi are resistant to most antifungal agents, including most of the azoles. For this reason, the drugs of choice to treat such infections are limited. The exploration of ergosterol biosynthesis is therefore of fundamental importance to understand the azole resistance of mucormycosis-causing fungi and to develop possible new control strategies. Characterization of sterol 24-C-methyltransferase demonstrated its role in the azole resistance and virulence of M. lusitanicus. Moreover, our experiments suggest that there are at least four alternative pathways for the biosynthesis of sterols in Mucor. Switching between pathways may contribute to the maintenance of azole resistance.
Keywords: CRISPR-Cas9; azole resistance; eburicol; erg6 gene; ergosterol; mucormycosis; zymosterol.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Alternative ergosterol biosynthetic pathways confer antifungal drug resistance in the human pathogens within the Mucor species complex.mBio. 2024 Aug 14;15(8):e0166124. doi: 10.1128/mbio.01661-24. Epub 2024 Jul 9. mBio. 2024. PMID: 38980037 Free PMC article.
-
Alternative ergosterol biosynthetic pathways confer antifungal drug resistance in the human pathogens within the Mucor species complex.bioRxiv [Preprint]. 2024 Jun 1:2023.12.01.569667. doi: 10.1101/2023.12.01.569667. bioRxiv. 2024. Update in: mBio. 2024 Aug 14;15(8):e0166124. doi: 10.1128/mbio.01661-24. PMID: 38076934 Free PMC article. Updated. Preprint.
-
Coordinated Regulation of Membrane Homeostasis and Drug Accumulation by Novel Kinase STK-17 in Response to Antifungal Azole Treatment.Microbiol Spectr. 2022 Feb 23;10(1):e0012722. doi: 10.1128/spectrum.00127-22. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196787 Free PMC article.
-
Molecular basis of resistance to azole antifungals.Trends Mol Med. 2002 Feb;8(2):76-81. doi: 10.1016/s1471-4914(02)02280-3. Trends Mol Med. 2002. PMID: 11815273 Review.
-
[Azole resistance in Candida spp].Nihon Ishinkin Gakkai Zasshi. 2003;44(2):87-92. doi: 10.3314/jjmm.44.87. Nihon Ishinkin Gakkai Zasshi. 2003. PMID: 12748589 Review. Japanese.
Cited by
-
Molecular mechanisms that govern infection and antifungal resistance in Mucorales.Microbiol Mol Biol Rev. 2024 Mar 27;88(1):e0018822. doi: 10.1128/mmbr.00188-22. Epub 2024 Mar 6. Microbiol Mol Biol Rev. 2024. PMID: 38445820 Free PMC article. Review.
-
Alternative ergosterol biosynthetic pathways confer antifungal drug resistance in the human pathogens within the Mucor species complex.mBio. 2024 Aug 14;15(8):e0166124. doi: 10.1128/mbio.01661-24. Epub 2024 Jul 9. mBio. 2024. PMID: 38980037 Free PMC article.
-
Role of the putative sit1 gene in normal germination of spores and virulence of the Mucor lusitanicus.Microb Cell. 2025 Aug 12;12:195-209. doi: 10.15698/mic2025.08.856. eCollection 2025. Microb Cell. 2025. PMID: 40873928 Free PMC article.
-
The sterol C-24 methyltransferase encoding gene, erg6, is essential for viability of Aspergillus species.Nat Commun. 2024 May 20;15(1):4261. doi: 10.1038/s41467-024-48767-3. Nat Commun. 2024. PMID: 38769341 Free PMC article.
-
Functional characterization of two survival factor 1 genes in Mucor lusitanicus.Microbiol Spectr. 2024 Oct 3;12(10):e0110324. doi: 10.1128/spectrum.01103-24. Epub 2024 Aug 27. Microbiol Spectr. 2024. PMID: 39189757 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

