Large Multicountry Outbreak of Invasive Listeriosis by a Listeria monocytogenes ST394 Clone Linked to Smoked Rainbow Trout, 2020 to 2021
- PMID: 37036341
- PMCID: PMC10269727
- DOI: 10.1128/spectrum.03520-22
Large Multicountry Outbreak of Invasive Listeriosis by a Listeria monocytogenes ST394 Clone Linked to Smoked Rainbow Trout, 2020 to 2021
Abstract
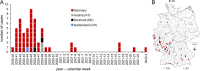
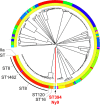
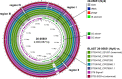
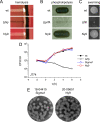
Whole-genome sequencing (WGS) has revolutionized surveillance of infectious diseases. Disease outbreaks can now be detected with high precision, and correct attribution of infection sources has been improved. Listeriosis, caused by the bacterium Listeria monocytogenes, is a foodborne disease with a high case fatality rate and a large proportion of outbreak-related cases. Timely recognition of listeriosis outbreaks and precise allocation of food sources are important to prevent further infections and to promote public health. We report the WGS-based identification of a large multinational listeriosis outbreak with 55 cases that affected Germany, Austria, Denmark, and Switzerland during 2020 and 2021. Clinical isolates formed a highly clonal cluster (called Ny9) based on core genome multilocus sequence typing (cgMLST). Routine and ad hoc investigations of food samples identified L. monocytogenes isolates from smoked rainbow trout filets from a Danish producer grouping with the Ny9 cluster. Patient interviews confirmed consumption of rainbow trout as the most likely infection source. The Ny9 cluster was caused by a MLST sequence type (ST) ST394 clone belonging to molecular serogroup IIa, forming a distinct clade within molecular serogroup IIa strains. Analysis of the Ny9 genome revealed clpY, dgcB, and recQ inactivating mutations, but phenotypic characterization of several virulence-associated traits of a representative Ny9 isolate showed that the outbreak strain had the same pathogenic potential as other serogroup IIa strains. Our report demonstrates that international food trade can cause multicountry outbreaks that necessitate cross-border outbreak collaboration. It also corroborates the relevance of ready-to-eat smoked fish products as causes for listeriosis. IMPORTANCE Listeriosis is a severe infectious disease in humans and characterized by an exceptionally high case fatality rate. The disease is transmitted through consumption of food contaminated by the bacterium Listeria monocytogenes. Outbreaks of listeriosis often occur but can be recognized and stopped through implementation of whole-genome sequencing-based pathogen surveillance systems. We here describe the detection and management of a large listeriosis outbreak in Germany and three neighboring countries. This outbreak was caused by rainbow trout filet, which was contaminated by a L. monocytogenes clone belonging to sequence type ST394. This work further expands our knowledge on the genetic diversity and transmission routes of an important foodborne pathogen.
Keywords: Europe; Ny; Ny9; ST394; cgMLST; epidemiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Retrospective investigation of listeriosis outbreaks in small ruminants using different analytical approaches for whole genome sequencing-based typing of Listeria monocytogenes.Infect Genet Evol. 2020 Jan;77:104047. doi: 10.1016/j.meegid.2019.104047. Epub 2019 Oct 17. Infect Genet Evol. 2020. PMID: 31629888
-
Rapid detection of the source of a Listeria monocytogenes outbreak in Switzerland through routine interviewing of patients and whole-genome sequencing.Swiss Med Wkly. 2024 May 3;154:3745. doi: 10.57187/s.3745. Swiss Med Wkly. 2024. PMID: 38701492
-
Core Genome Multilocus Sequence Typing for Identification of Globally Distributed Clonal Groups and Differentiation of Outbreak Strains of Listeria monocytogenes.Appl Environ Microbiol. 2016 Sep 30;82(20):6258-6272. doi: 10.1128/AEM.01532-16. Print 2016 Oct 15. Appl Environ Microbiol. 2016. PMID: 27520821 Free PMC article.
-
Lessons from an outbreak of listeriosis related to vacuum-packed gravad and cold-smoked fish.Int J Food Microbiol. 2000 Dec 20;62(3):173-5. doi: 10.1016/s0168-1605(00)00332-9. Int J Food Microbiol. 2000. PMID: 11156259 Review.
-
Wholegenome sequencing as the gold standard approach for control of Listeria monocytogenes in the food chain.J Food Prot. 2023 Jan;86(1):100003. doi: 10.1016/j.jfp.2022.10.002. Epub 2022 Dec 16. J Food Prot. 2023. PMID: 36916580 Review.
Cited by
-
The European Union One Health 2023 Zoonoses report.EFSA J. 2024 Dec 10;22(12):e9106. doi: 10.2903/j.efsa.2024.9106. eCollection 2024 Dec. EFSA J. 2024. PMID: 39659847 Free PMC article.
-
ClpP2 proteasomes and SpxA1 determine Listeria monocytogenes tartrolon B hyper-resistance.PLoS Genet. 2025 Apr 4;21(4):e1011621. doi: 10.1371/journal.pgen.1011621. eCollection 2025 Apr. PLoS Genet. 2025. PMID: 40184427 Free PMC article.
-
Current methodologies available to evaluate the virulence potential among Listeria monocytogenes clonal complexes.Front Microbiol. 2024 Oct 10;15:1425437. doi: 10.3389/fmicb.2024.1425437. eCollection 2024. Front Microbiol. 2024. PMID: 39493856 Free PMC article. Review.
-
Impact of Nisin on Proliferation of Background Microbiota, Pressure-Stressed and Wild-Type Listeria monocytogenes, and Listeria innocua During a Real-Time Shelf-Life Study.Microorganisms. 2025 Mar 15;13(3):668. doi: 10.3390/microorganisms13030668. Microorganisms. 2025. PMID: 40142560 Free PMC article.
-
From Traditional Typing to Genomic Precision: Whole-Genome Sequencing of Listeria monocytogenes Isolated from Refrigerated Foods in Chile.Foods. 2025 Jan 16;14(2):290. doi: 10.3390/foods14020290. Foods. 2025. PMID: 39856956 Free PMC article.
References
-
- Charlier C, Perrodeau E, Leclercq A, Cazenave B, Pilmis B, Henry B, Lopes A, Maury MM, Moura A, Goffinet F, Dieye HB, Thouvenot P, Ungeheuer MN, Tourdjman M, Goulet V, de Valk H, Lortholary O, Ravaud P, Lecuit M, Group M. 2017. Clinical features and prognostic factors of listeriosis: the MONALISA national prospective cohort study. Lancet Infect Dis 17:510–519. doi:10.1016/S1473-3099(16)30521-7. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

