FOXK1 regulates Wnt signalling to promote cardiogenesis
- PMID: 37036809
- PMCID: PMC10325700
- DOI: 10.1093/cvr/cvad054
FOXK1 regulates Wnt signalling to promote cardiogenesis
Abstract
Aims: Congenital heart disease (CHD) is the most common genetic birth defect, which has considerable morbidity and mortality. We focused on deciphering key regulators that govern cardiac progenitors and cardiogenesis. FOXK1 is a forkhead/winged helix transcription factor known to regulate cell cycle kinetics and is restricted to mesodermal progenitors, somites, and heart. In the present study, we define an essential role for FOXK1 during cardiovascular development.
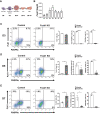
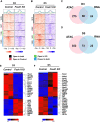
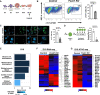
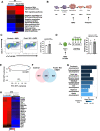
Methods and results: We used the mouse embryoid body system to differentiate control and Foxk1 KO embryonic stem cells into mesodermal, cardiac progenitor cells and mature cardiac cells. Using flow cytometry, immunohistochemistry, cardiac beating, transcriptional and chromatin immunoprecipitation quantitative polymerase chain reaction assays, bulk RNA sequencing (RNAseq) and assay for transposase-accessible chromatin using sequencing (ATACseq) analyses, FOXK1 was observed to be an important regulator of cardiogenesis. Flow cytometry analyses revealed perturbed cardiogenesis in Foxk1 KO embryoid bodies (EBs). Bulk RNAseq analysis at two developmental stages showed a significant reduction of the cardiac molecular program in Foxk1 KO EBs compared to the control EBs. ATACseq analysis during EB differentiation demonstrated that the chromatin landscape nearby known important regulators of cardiogenesis was significantly relaxed in control EBs compared to Foxk1 KO EBs. Furthermore, we demonstrated that in the absence of FOXK1, cardiac differentiation was markedly impaired by assaying for cardiac Troponin T expression and cardiac contractility. We demonstrate that FOXK1 is an important regulator of cardiogenesis by repressing the Wnt/β-catenin signalling pathway and thereby promoting differentiation.
Conclusion: These results identify FOXK1 as an essential transcriptional and epigenetic regulator of cardiovascular development. Mechanistically, FOXK1 represses Wnt signalling to promote the development of cardiac progenitor cells.
Keywords: Forkhead factors; Cardiovascular development; Foxk1; Wnt signalling.
© The Author(s) 2023. Published by Oxford University Press on behalf of the European Society of Cardiology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest: None declared.
Figures


Similar articles
-
Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11.Proc Natl Acad Sci U S A. 2009 Oct 6;106(40):16978-83. doi: 10.1073/pnas.0905550106. Epub 2009 Sep 23. Proc Natl Acad Sci U S A. 2009. PMID: 19805103 Free PMC article.
-
The four and a half LIM-domain 2 controls early cardiac cell commitment and expansion via regulating β-catenin-dependent transcription.Stem Cells. 2013 May;31(5):928-40. doi: 10.1002/stem.1332. Stem Cells. 2013. PMID: 23341242 Free PMC article.
-
Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells.Proc Natl Acad Sci U S A. 2007 Jun 5;104(23):9685-90. doi: 10.1073/pnas.0702859104. Epub 2007 May 23. Proc Natl Acad Sci U S A. 2007. PMID: 17522258 Free PMC article.
-
Wnt Signaling: The double-edged sword diminishing the potential of stem cell therapy in congenital heart disease.Life Sci. 2019 Dec 15;239:116937. doi: 10.1016/j.lfs.2019.116937. Epub 2019 Oct 17. Life Sci. 2019. PMID: 31629761 Review.
-
Wnt/ß-catenin signalling and the dynamics of fate decisions in early mouse embryos and embryonic stem (ES) cells.Semin Cell Dev Biol. 2015 Dec;47-48:101-9. doi: 10.1016/j.semcdb.2015.08.011. Epub 2015 Aug 29. Semin Cell Dev Biol. 2015. PMID: 26321498 Free PMC article. Review.
Cited by
-
Cognitive Impairment in Multiple Sclerosis.Bioengineering (Basel). 2023 Jul 23;10(7):871. doi: 10.3390/bioengineering10070871. Bioengineering (Basel). 2023. PMID: 37508898 Free PMC article. Review.
-
ETV2 transcriptionally activates Rig1 gene expression and promotes reprogramming of the endothelial lineage.Sci Rep. 2024 Nov 19;14(1):28688. doi: 10.1038/s41598-024-78115-w. Sci Rep. 2024. PMID: 39562637 Free PMC article.
-
The impact of aging and oxidative stress in metabolic and nervous system disorders: programmed cell death and molecular signal transduction crosstalk.Front Immunol. 2023 Nov 8;14:1273570. doi: 10.3389/fimmu.2023.1273570. eCollection 2023. Front Immunol. 2023. PMID: 38022638 Free PMC article. Review.
-
Foxk1 and Foxk2 promote cardiomyocyte proliferation and heart regeneration.Nat Commun. 2025 Mar 24;16(1):2877. doi: 10.1038/s41467-025-57996-z. Nat Commun. 2025. PMID: 40128196 Free PMC article.
-
Foxk1 promotes bone formation through inducing aerobic glycolysis.Cell Death Differ. 2024 Dec;31(12):1650-1663. doi: 10.1038/s41418-024-01371-w. Epub 2024 Sep 5. Cell Death Differ. 2024. PMID: 39232134
References
-
- Hoffman JIE. Incidence of congenital heart disease: I. Postnatal incidence. Pediatr Cardiol 1995;16:103–113. - PubMed
-
- Bruneau BG. The developmental genetics of congenital heart disease. Nature 2008;451:943–948. - PubMed
-
- Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol 2012;74:41–68. - PubMed
-
- Tam PPL, Behringer RR. Mouse gastrulation: the formation of a mammalian body plan. Mech Dev 1997;68:3–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

