MmuPV1 E7's interaction with PTPN14 delays Epithelial differentiation and contributes to virus-induced skin disease
- PMID: 37036883
- PMCID: PMC10085053
- DOI: 10.1371/journal.ppat.1011215
MmuPV1 E7's interaction with PTPN14 delays Epithelial differentiation and contributes to virus-induced skin disease
Abstract
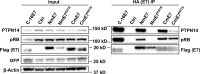
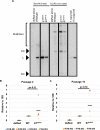
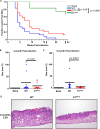
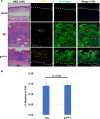
Human papillomaviruses (HPVs) contribute to approximately 5% of all human cancers. Species-specific barriers limit the ability to study HPV pathogenesis in animal models. Murine papillomavirus (MmuPV1) provides a powerful tool to study the roles of papillomavirus genes in pathogenesis arising from a natural infection. We previously identified Protein Tyrosine Phosphatase Non-Receptor Type 14 (PTPN14), a tumor suppressor targeted by HPV E7 proteins, as a putative cellular target of MmuPV1 E7. Here, we confirmed the MmuPV1 E7-PTPN14 interaction. Based on the published structure of the HPV18 E7/PTPN14 complex, we generated a MmuPV1 E7 mutant, E7K81S, that was defective for binding PTPN14. Wild-type (WT) and E7K81S mutant viral genomes replicated as extrachromosomal circular DNAs to comparable levels in mouse keratinocytes. E7K81S mutant virus (E7K81S MmuPV1) was generated and used to infect FoxN/Nude mice. E7K81S MmuPV1 caused neoplastic lesions at a frequency similar to that of WT MmuPV1, but the lesions arose later and were smaller than WT-induced lesions. The E7K81S MmuPV1-induced lesions also had a trend towards a less severe grade of neoplastic disease. In the lesions, E7K81S MmuPV1 supported the late (productive) stage of the viral life cycle and promoted E2F activity and cellular DNA synthesis in suprabasal epithelial cells to similar degrees as WT MmuPV1. There was a similar frequency of lateral spread of infections among mice infected with E7K81S or WT MmuPV1. Compared to WT MmuPV1-induced lesions, E7K81S MmuPV1-induced lesions had a significant expansion of cells expressing differentiation markers, Keratin 10 and Involucrin. We conclude that an intact PTPN14 binding site is necessary for MmuPV1 E7's ability to contribute to papillomavirus-induced pathogenesis and this correlates with MmuPV1 E7 causing a delay in epithelial differentiation, which is a hallmark of papillomavirus-induced neoplasia.
Copyright: © 2023 Romero-Masters et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
The Mus musculus Papillomavirus Type 1 E7 Protein Binds to the Retinoblastoma Tumor Suppressor: Implications for Viral Pathogenesis.mBio. 2021 Aug 31;12(4):e0227721. doi: 10.1128/mBio.02277-21. Epub 2021 Aug 31. mBio. 2021. PMID: 34465025 Free PMC article.
-
A Conserved Amino Acid in the C Terminus of Human Papillomavirus E7 Mediates Binding to PTPN14 and Repression of Epithelial Differentiation.J Virol. 2020 Aug 17;94(17):e01024-20. doi: 10.1128/JVI.01024-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32581101 Free PMC article.
-
Structural basis for recognition of the tumor suppressor protein PTPN14 by the oncoprotein E7 of human papillomavirus.PLoS Biol. 2019 Jul 19;17(7):e3000367. doi: 10.1371/journal.pbio.3000367. eCollection 2019 Jul. PLoS Biol. 2019. PMID: 31323018 Free PMC article.
-
Molecular Mechanisms of MmuPV1 E6 and E7 and Implications for Human Disease.Viruses. 2022 Sep 28;14(10):2138. doi: 10.3390/v14102138. Viruses. 2022. PMID: 36298698 Free PMC article. Review.
-
Papillomavirus E6 and E7 proteins and their cellular targets.Front Biosci. 2008 Jan 1;13:1003-17. doi: 10.2741/2739. Front Biosci. 2008. PMID: 17981607 Review.
Cited by
-
mSphere of Influence: MmuPV1-a dual tropic papillomavirus, red herring, or novel insight into HPV pathogenesis.mSphere. 2024 Jul 30;9(7):e0017724. doi: 10.1128/msphere.00177-24. Epub 2024 Jun 26. mSphere. 2024. PMID: 38920397 Free PMC article.
-
Monitoring mouse papillomavirus-associated cancer development using longitudinal Pap smear screening.mBio. 2024 Aug 14;15(8):e0142024. doi: 10.1128/mbio.01420-24. Epub 2024 Jul 16. mBio. 2024. PMID: 39012151 Free PMC article.
-
Human papillomaviruses: Knowns, mysteries, and unchartered territories.J Med Virol. 2023 Oct;95(10):e29191. doi: 10.1002/jmv.29191. J Med Virol. 2023. PMID: 37861365 Free PMC article. Review.
-
The Larynx is Protected from Secondary and Vertical Papillomavirus Infection in Immunocompetent Mice.Laryngoscope. 2024 May;134(5):2322-2330. doi: 10.1002/lary.31228. Epub 2023 Dec 12. Laryngoscope. 2024. PMID: 38084790 Free PMC article.
References
-
- Liu L. Fields Virology, 6th Edition. Clinical Infectious Diseases. 2014;59: 613. doi: 10.1093/cid/ciu346 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

