Identification of Spatial Proteomic Signatures of Colon Tumor Metastasis: A Digital Spatial Profiling Approach
- PMID: 37037284
- PMCID: PMC10284031
- DOI: 10.1016/j.ajpath.2023.02.020
Identification of Spatial Proteomic Signatures of Colon Tumor Metastasis: A Digital Spatial Profiling Approach
Abstract
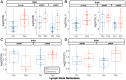
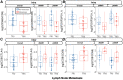
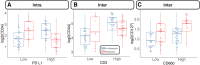
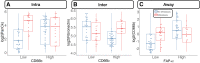
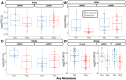
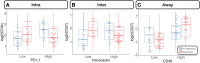
Over 150,000 Americans are diagnosed with colorectal cancer (CRC) every year, and annually >50,000 individuals are estimated to die of CRC, necessitating improvements in screening, prognostication, disease management, and therapeutic options. CRC tumors are removed en bloc with surrounding vasculature and lymphatics. Examination of regional lymph nodes at the time of surgical resection is essential for prognostication. Developing alternative approaches to indirectly assess recurrence risk would have utility in cases where lymph node yield is incomplete or inadequate. Spatially dependent, immune cell-specific (eg, tumor-infiltrating lymphocytes), proteomic, and transcriptomic expression patterns inside and around the tumor-the tumor immune microenvironment-can predict nodal/distant metastasis and probe the coordinated immune response from the primary tumor site. The comprehensive characterization of tumor-infiltrating lymphocytes and other immune infiltrates is possible using highly multiplexed spatial omics technologies, such as the GeoMX Digital Spatial Profiler. In this study, machine learning and differential co-expression analyses helped identify biomarkers from Digital Spatial Profiler-assayed protein expression patterns inside, at the invasive margin, and away from the tumor, associated with extracellular matrix remodeling (eg, granzyme B and fibronectin), immune suppression (eg, forkhead box P3), exhaustion and cytotoxicity (eg, CD8), Programmed death ligand 1-expressing dendritic cells, and neutrophil proliferation, among other concomitant alterations. Further investigation of these biomarkers may reveal independent risk factors of CRC metastasis that can be formulated into low-cost, widely available assays.
Copyright © 2023 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures




















References
-
- Kasi P.M., Shahjehan F., Cochuyt J.J., Li Z., Colibaseanu D.T., Merchea A. Rising proportion of young individuals with rectal and colon cancer. Clin Colorectal Cancer. 2019;18:e87–e95. - PubMed
-
- Slattery M.L. Diet, lifestyle, and colon cancer. Semin Gastrointest Dis. 2000;11:142–146. - PubMed
-
- Siegel R.L., Miller K.D., Goding Sauer A., Fedewa S.A., Butterly L.F., Anderson J.C., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

