Proteomic analysis reveals microvesicles containing NAMPT as mediators of radioresistance in glioma
- PMID: 37037593
- PMCID: PMC10087103
- DOI: 10.26508/lsa.202201680
Proteomic analysis reveals microvesicles containing NAMPT as mediators of radioresistance in glioma
Abstract
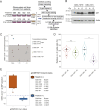
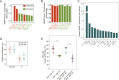
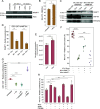
Tumor-initiating cells contained within the aggressive brain tumor glioma (glioma stem cells, GSCs) promote radioresistance and disease recurrence. However, mechanisms of resistance are not well understood. Herein, we show that the proteome-level regulation occurring upon radiation treatment of several patient-derived GSC lines predicts their resistance status, whereas glioma transcriptional subtypes do not. We identify a mechanism of radioresistance mediated by the transfer of the metabolic enzyme NAMPT to radiosensitive cells through microvesicles (NAMPT-high MVs) shed by resistant GSCs. NAMPT-high MVs rescue the proliferation of radiosensitive GSCs and fibroblasts upon irradiation, and upon treatment with a radiomimetic drug or low serum, and increase intracellular NAD(H) levels. Finally, we show that the presence of NAMPT within the MVs and its enzymatic activity in recipient cells are necessary to mediate these effects. Collectively, we demonstrate that the proteome of GSCs provides unique information as it predicts the ability of glioma to resist radiation treatment. Furthermore, we establish NAMPT transfer via MVs as a mechanism for rescuing the proliferation of radiosensitive cells upon irradiation.
© 2023 Panizza et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Alexa A, Rahnenfuhrer J (2021) topGO: Enrichment Analysis for Gene Ontology. (R package version 2.46.0). Bioconductor. 10.18129/B9.bioc.topGO - DOI
-
- Alhalabi OT, Fletcher MNC, Hielscher T, Kessler T, Lokumcu T, Baumgartner U, Wittmann E, Schlue S, Göttmann M, Rahman S, et al. (2022) A novel patient stratification strategy to enhance the therapeutic efficacy of dasatinib in glioblastoma. Neuro-Oncology 24: 39–51. 10.1093/NEUONC/NOAB158 - DOI - PMC - PubMed
-
- Audrito V, Serra S, Brusa D, Mazzola F, Arruga F, Vaisitti T, Coscia M, Maffei R, Rossi D, Wang T, et al. (2015) Extracellular nicotinamide phosphoribosyltransferase (NAMPT) promotes M2 macrophage polarization in chronic lymphocytic leukemia. Blood 125: 111–123. 10.1182/BLOOD-2014-07-589069 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
