Prior anti-CTLA-4 therapy impacts molecular characteristics associated with anti-PD-1 response in advanced melanoma
- PMID: 37037616
- PMCID: PMC10187051
- DOI: 10.1016/j.ccell.2023.03.010
Prior anti-CTLA-4 therapy impacts molecular characteristics associated with anti-PD-1 response in advanced melanoma
Abstract
Immune checkpoint inhibitors (ICIs), including CTLA-4- and PD-1-blocking antibodies, can have profound effects on tumor immune cell infiltration that have not been consistent in biopsy series reported to date. Here, we analyze seven molecular datasets of samples from patients with advanced melanoma (N = 514) treated with ICI agents to investigate clinical, genomic, and transcriptomic features of anti-PD-1 response in cutaneous melanoma. We find that prior anti-CTLA-4 therapy is associated with differences in genomic, individual gene, and gene signatures in anti-PD-1 responders. Anti-CTLA-4-experienced melanoma tumors that respond to PD-1 blockade exhibit increased tumor mutational burden, inflammatory signatures, and altered cell cycle processes compared with anti-CTLA-4-naive tumors or anti-CTLA-4-experienced, anti-PD-1-nonresponsive melanoma tumors. We report a harmonized, aggregate resource and suggest that prior CTLA-4 blockade therapy is associated with marked differences in the tumor microenvironment that impact the predictive features of PD-1 blockade therapy response.
Keywords: immune checkpoint blockade; immunotherapy; melanoma; meta-analysis.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.M.C. is a shareholder in Geneoscopy LLC and received consulting fees from PACT Pharma, Tango Therapeutics, Geneoscopy LLC, Flagship Labs 81 LLC, and the Rare Cancer Research Foundation. M.A. received consulting fees from PICI. J.S.W. received consulting fees from Merck, Genentech, AstraZeneca, GlaxoSmithKline, Novartis, Nektar, Celldex, Incyte, Biond, ImCheck, Sellas, Evaxion, and EMD Serono; is or has been a member of the scientific advisory board for Bristol-Myers Squibb (BMS), CytoMx, Incyte, ImCheck, Biond, Sellas, Instil Bio, OncoC4 and Neximmune; and holds equity in Biond, Evaxion, Instil Bio, and Neximmune. J.S.W. has been named on patents filed by Moffitt Cancer Center (ipilimumab biomarker and TIL growth method) and Biosedix (PD-1). NYU has received research support from BMS, Merck, GlaxoSmithKline, Moderna, Pfizer, Novartis, and AstraZeneca. J.D.W. received consulting fees from Apricity, CellCarta, Ascentage Pharma, AstraZeneca, Astellas, Bicara Therapeutics, Boehringer Ingelheim, BMS, Dragonfly, Georgiamune, Imvaq, Larkspur, Maverick Therapeutics, Psioxus, Recepta, Tizona, and Sellas; has received research/grant support from BMS and Sephora; and has equity in Apricity, Arsenal IO, Ascentage, Beigene, Imvaq, Linnaeus, Georgiamune, Maverick, Tizona Pharmaceuticals, and Trieza. J.L. has worked in a consulting/advisory role for iOnctura, Apple Tree, Merck, BMS, Eisai, Debipharm, and Incyte; received honoraria from AstraZeneca, BMS, Eisai, EUSA Pharma, GlaxoSmithKline, Incyte, Ipsen, Merck, touchEXPERTS, Royal College of Physicians, Cambridge Healthcare Research, Royal College of General Practitioners, VJOncology, Agence Unik, Merck Sharp & Dohme, Novartis, Aptitude, Pierre Fabre, Pfizer, Roche, Seagen, Inselgruppe, eCancer, Ultimovacs, Calithera, and Goldman Sachs; and received research/grant support from Achilles Therapeutics, BMS, Immunocore, Aveo, Pharmacyclics, MSD, Nektar Therapeutics, Covance, Novartis, Pfizer, and Roche. F.S.H. received research support from the NCI of the NIH, BMS, Novartis, and Genentech; royalties or licenses from BMS and Novartis; consulting fees from BMS, EMD Serono, Surface, Sanofi, Genentech, Gossamer, Trillium, Immunocore, Merck, Novartis, Compass Therapeutics, Pieris, Bioentre, Iovance, Catalym, and Amgen; patents for methods for treating MHC class I polypeptide-related sequence A disorders (20100111973, pending, with royalties paid), tumor antigens and uses thereof (7250291, issued), angiopoiten-2 biomarkers predictive of anti-immune checkpoint response (20170248603, pending), compositions and methods for the identification, assessment, prevention, and treatment of melanoma using PD-L1 isoforms (20160340407, pending), therapeutic peptides (20160046716, 20140004112, 20170022275, and 20170008962, all pending, and 9402905, issued), methods of using pembrolizumab and trebananib (pending), vaccine compositions and methods for restoring NKG2D pathway function against cancers (10279021, issued), antibodies that bind to MHC class I polypeptide-related sequence A (10106611, issued), and anti-galectin antibody biomarkers predictive of anti-immune checkpoint and anti-angiogenesis responses (20170343552, pending); data safety monitoring board and advisory board participation for Aduro and Checkpoint Therapeutics; scientific advisory board leadership for Bicara and Apricity; and stock options in Checkpoint Therapeutics, Pionyr, Apricity, and Bicara. S.B., L.S., D.T., and T.T. are employees of BMS. C.N.S. received consulting fees from the Rare Cancer Research Foundation. D.K.W. is an employee, founder, and equity holder at Santa Ana Bio; holds equity in Immunai, and has received consulting fees from Rubius Therapeutics, DeepMind, Illumina, and Guardant Health. A.R. has received honoraria from consulting with Amgen, BMS, Chugai, Genentech, Merck, Novartis, Roche, and Sanofi; is or has been a member of the scientific advisory board and holds stock in Advaxis, Arcus Biosciences, Bioncotech Therapeutics, Compugen, CytomX, Five Prim, FLX-Bio, ImaginAb, Isoplexis, Kite-Gilead, Lutris Pharma, Merus, PACT Pharma, Rgenix, and Tango Therapeutics; and has received research funding from Agilent Technologies and BMS through Stand Up to Cancer.
Figures

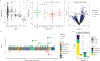




References
-
- Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, et al. (2015). Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 372, 2521–2532. - PubMed
-
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob J-J, Rutkowski P, Lao CD, Cowey CL, Schadendorf D, Wagstaff J, Dummer R, et al. (2019). Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 381, 1535–1546. - PubMed
-
- Center for Drug Evaluation, and Research FDA grants accelerated approval to dostarlimab-gxly for dMMR advanced solid tumors. U.S. Food and Drug Administration. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grant....

