Uropathogenic Escherichia coli infection-induced epithelial trained immunity impacts urinary tract disease outcome
- PMID: 37037942
- PMCID: PMC10159856
- DOI: 10.1038/s41564-023-01346-6
Uropathogenic Escherichia coli infection-induced epithelial trained immunity impacts urinary tract disease outcome
Abstract
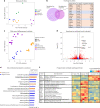
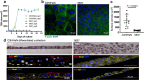
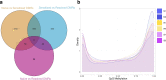
Previous urinary tract infections (UTIs) can predispose one to future infections; however, the underlying mechanisms affecting recurrence are poorly understood. We previously found that UTIs in mice cause differential bladder epithelial (urothelial) remodelling, depending on disease outcome, that impacts susceptibility to recurrent UTI. Here we compared urothelial stem cell (USC) lines isolated from mice with a history of either resolved or chronic uropathogenic Escherichia coli (UPEC) infection, elucidating evidence of molecular imprinting that involved epigenetic changes, including differences in chromatin accessibility, DNA methylation and histone modification. Epigenetic marks in USCs from chronically infected mice enhanced caspase-1-mediated cell death upon UPEC infection, promoting bacterial clearance. Increased Ptgs2os2 expression also occurred, potentially contributing to sustained cyclooxygenase-2 expression, bladder inflammation and mucosal wounding-responses associated with severe recurrent cystitis. Thus, UPEC infection acts as an epi-mutagen reprogramming the urothelial epigenome, leading to urothelial-intrinsic remodelling and training of the innate response to subsequent infection.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












Comment in
-
Uropathogenic E. coli creates a memory.Nat Rev Microbiol. 2023 Jun;21(6):345. doi: 10.1038/s41579-023-00898-y. Nat Rev Microbiol. 2023. PMID: 37059876 No abstract available.
-
Epigenetic modifications influence urinary tract infection outcome.Nat Microbiol. 2023 May;8(5):764-766. doi: 10.1038/s41564-023-01371-5. Nat Microbiol. 2023. PMID: 37069403 No abstract available.
-
Uro-Science.J Urol. 2024 Jan;211(1):194-195. doi: 10.1097/JU.0000000000003710. Epub 2023 Oct 20. J Urol. 2024. PMID: 37861082 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

