PD-L1 blockade in combination with carboplatin as immune induction in metastatic lobular breast cancer: the GELATO trial
- PMID: 37038006
- PMCID: PMC10132987
- DOI: 10.1038/s43018-023-00542-x
PD-L1 blockade in combination with carboplatin as immune induction in metastatic lobular breast cancer: the GELATO trial
Abstract
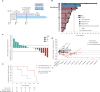
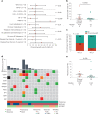
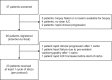
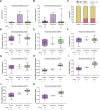
Invasive lobular breast cancer (ILC) is the second most common histological breast cancer subtype, but ILC-specific trials are lacking. Translational research revealed an immune-related ILC subset, and in mouse ILC models, synergy between immune checkpoint blockade and platinum was observed. In the phase II GELATO trial ( NCT03147040 ), patients with metastatic ILC were treated with weekly carboplatin (area under the curve 1.5 mg ml-1 min-1) as immune induction for 12 weeks and atezolizumab (PD-L1 blockade; triweekly) from the third week until progression. Four of 23 evaluable patients had a partial response (17%), and 2 had stable disease, resulting in a clinical benefit rate of 26%. From these six patients, four had triple-negative ILC (TN-ILC). We observed higher CD8+ T cell infiltration, immune checkpoint expression and exhausted T cells after treatment. With this GELATO trial, we show that ILC-specific clinical trials are feasible and demonstrate promising antitumor activity of atezolizumab with carboplatin, particularly for TN-ILC, and provide insights for the design of highly needed ILC-specific trials.
© 2023. The Author(s).
Conflict of interest statement
L.V., O.I.I., S.B., M.C., N.A.M.B., E.C., H.G., K.S., C.E.L., I.K., I.A.M.M., M.d.M., J.J.L.v.G., J.B., M.G.J.v.D. and A.J. have no competing interests to declare. H.M.H. reports consultancy fees from Roche Diagnostics paid to the institute and is an advisor for SlideScore and Ellogon from Roche Diagnostics, outside the submitted work. M.d.B. received funding from Roche, AstraZeneca, Novartis, Pfizer, Eisai and Eli Lilly, outside the submitted work. R.S. reports non-financial support from Merck and Bristol Myers Squibb (BMS), research support from Merck, Puma Biotechnology and Roche and personal fees from Roche, BMS and Exact Sciences for advisory boards, all outside the scope of this paper. G.S.S. reports research funding to the institute from Merck, Agendia, AstraZeneca, Roche and Novartis and a consulting role for Novartis, Seattle Genetics and Biovica, outside the submitted work. K.E.d.V. reports research funding from Roche and is a consultant for Macomics, outside the scope of this work. T.N.S. is a consultant for Third Rock Ventures and is a stockholder and advisor in Allogene Therapeutics, Asher Bio, Celsius, Merus, Scenic Biotech and Neogene Therapeutics, outside this work. C.U.B. has received research grants from Novartis, BMS and NanoString, is a paid advisory board member for BMS, MSD, Roche, Novartis, GlaxoSmithKline, AstraZeneca, Pfizer, Lilly, GenMab and Pierre Fabre and holds ownership interest in Uniti Card, Neon Therapeutics and Forty Seven, all outside this submitted work. L.F.A.W. reports funding to the institute from Genmab BV. V.C.G.T.-H. reports research funding to the institute from Roche, Eisai, Pfizer, Novartis, Lilly, Daiichi Sankyo/AstraZeneca and Gilead Sciences, a consulting role from Pfizer, Lilly, Accord Healthcare and Novartis and honoraria from Novartis, Roche, Lilly and AstraZeneca, all outside this submitted work. C.P.S. has received research funding to the institute from Pfizer, Roche, Genentech, SNS Oncology, G1 Therapeutics, Abbvie, Synthon and CytoMx Therapeutics, outside this work. S.C.L. reports research funding to the institute from Roche/Genentech, AstraZeneca, BMS, Tesaro, Merck, Immunomedics, Eurocept Pharmaceuticals, Agendia and Novartis and a consulting role and travel grant from Daiichi Sankyo, outside this work. M.K. reports funding to the institute from BMS, Roche and AstraZeneca/MedImmune and an advisory role for BMS, Roche, MSD and Daiichi Sankyo, outside the submitted work.
Figures







Similar articles
-
IMpassion132 Phase III trial: atezolizumab and chemotherapy in early relapsing metastatic triple-negative breast cancer.Future Oncol. 2019 Jun;15(17):1951-1961. doi: 10.2217/fon-2019-0059. Epub 2019 Apr 12. Future Oncol. 2019. PMID: 30977385
-
Pathologic complete response (pCR) to neoadjuvant treatment with or without atezolizumab in triple-negative, early high-risk and locally advanced breast cancer: NeoTRIP Michelangelo randomized study.Ann Oncol. 2022 May;33(5):534-543. doi: 10.1016/j.annonc.2022.02.004. Epub 2022 Feb 17. Ann Oncol. 2022. PMID: 35182721 Clinical Trial.
-
Triple-negative breast lobular carcinoma: a luminal androgen receptor carcinoma with specific ESRRA mutations.Mod Pathol. 2021 Jul;34(7):1282-1296. doi: 10.1038/s41379-021-00742-9. Epub 2021 Mar 22. Mod Pathol. 2021. PMID: 33753865 Free PMC article.
-
ROSALINE: a phase II, neoadjuvant study targeting ROS1 in combination with endocrine therapy in invasive lobular carcinoma of the breast.Future Oncol. 2022 Jul;18(22):2383-2392. doi: 10.2217/fon-2022-0358. Epub 2022 Jun 13. Future Oncol. 2022. PMID: 35695563 Review.
-
Biomarkers of Immune Checkpoint Blockade Response in Triple-Negative Breast Cancer.Curr Treat Options Oncol. 2021 Mar 20;22(5):38. doi: 10.1007/s11864-021-00833-4. Curr Treat Options Oncol. 2021. PMID: 33743085 Review.
Cited by
-
The immunopeptidome of colon cancer cells treated with topoisomerase inhibiting drug reveals differential as well as common endogenous protein sampling and display of MHC I-associated peptides.Mol Cell Oncol. 2025 Mar 3;12(1):2471640. doi: 10.1080/23723556.2025.2471640. eCollection 2025. Mol Cell Oncol. 2025. PMID: 40051755 Free PMC article.
-
Optimized Modeling of Metastatic Triple-Negative Invasive Lobular Breast Carcinoma.Cancers (Basel). 2023 Jun 22;15(13):3299. doi: 10.3390/cancers15133299. Cancers (Basel). 2023. PMID: 37444409 Free PMC article.
-
Tumor Mutational Burden in Breast Cancer: Current Evidence, Challenges, and Opportunities.Cancers (Basel). 2023 Aug 7;15(15):3997. doi: 10.3390/cancers15153997. Cancers (Basel). 2023. PMID: 37568813 Free PMC article. Review.
-
Tumor Infiltrating Lymphocytes (TILS) and PD-L1 Expression in Breast Cancer: A Review of Current Evidence and Prognostic Implications from Pathologist's Perspective.Cancers (Basel). 2023 Sep 8;15(18):4479. doi: 10.3390/cancers15184479. Cancers (Basel). 2023. PMID: 37760449 Free PMC article. Review.
-
Reporting on invasive lobular breast cancer in clinical trials: a systematic review.NPJ Breast Cancer. 2024 Mar 20;10(1):23. doi: 10.1038/s41523-024-00627-5. NPJ Breast Cancer. 2024. PMID: 38509112 Free PMC article. Review.
References
-
- WHO Classification of Tumours Editorial Board. WHO Classification of Breast Tumours 5th edn, Vol. 2 (World Health Organization, 2019).
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

