Telomeric retrotransposons show propensity to form G-quadruplexes in various eukaryotic species
- PMID: 37038191
- PMCID: PMC10088271
- DOI: 10.1186/s13100-023-00291-9
Telomeric retrotransposons show propensity to form G-quadruplexes in various eukaryotic species
Abstract
Background: Canonical telomeres (telomerase-synthetised) are readily forming G-quadruplexes (G4) on the G-rich strand. However, there are examples of non-canonical telomeres among eukaryotes where telomeric tandem repeats are invaded by specific retrotransposons. Drosophila melanogaster represents an extreme example with telomeres composed solely by three retrotransposons-Het-A, TAHRE and TART (HTT). Even though non-canonical telomeres often show strand biased G-distribution, the evidence for the G4-forming potential is limited.
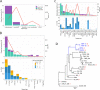
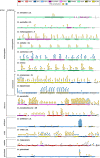
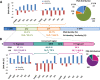
Results: Using circular dichroism spectroscopy and UV absorption melting assay we have verified in vitro G4-formation in the HTT elements of D. melanogaster. Namely 3 in Het-A, 8 in TART and 2 in TAHRE. All the G4s are asymmetrically distributed as in canonical telomeres. Bioinformatic analysis showed that asymmetric distribution of potential quadruplex sequences (PQS) is common in telomeric retrotransposons in other Drosophila species. Most of the PQS are located in the gag gene where PQS density correlates with higher DNA sequence conservation and codon selection favoring G4-forming potential. The importance of G4s in non-canonical telomeres is further supported by analysis of telomere-associated retrotransposons from various eukaryotic species including green algae, Diplomonadida, fungi, insects and vertebrates. Virtually all analyzed telomere-associated retrotransposons contained PQS, frequently with asymmetric strand distribution. Comparison with non-telomeric elements showed independent selection of PQS-rich elements from four distinct LINE clades.
Conclusion: Our findings of strand-biased G4-forming motifs in telomere-associated retrotransposons from various eukaryotic species support the G4-formation as one of the prerequisites for the recruitment of specific retrotransposons to chromosome ends and call for further experimental studies.
Keywords: Drosophila; G-quadruplex; Het-A; Retrotransposon; TAHRE; TART; Telomere.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

