Chemical Decrosslinking-Based Peptide Characterization of Formaldehyde-Fixed Rat Pancreas Using Fluorescence-Guided Single-Cell Mass Spectrometry
- PMID: 37040477
- PMCID: PMC10225058
- DOI: 10.1021/acs.analchem.3c00612
Chemical Decrosslinking-Based Peptide Characterization of Formaldehyde-Fixed Rat Pancreas Using Fluorescence-Guided Single-Cell Mass Spectrometry
Abstract
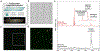
Approaches for the characterization of proteins/peptides in single cells of formaldehyde-fixed (FF) tissues via mass spectrometry (MS) are still under development. The lack of a general method for selectively eliminating formaldehyde-induced crosslinking is a major challenge. A workflow is shown for the high-throughput peptide profiling of single cells isolated from FF tissues, here the rodent pancreas, which possesses multiple peptide hormones from the islets of Langerhans. The heat treatment is enhanced by a collagen-selective multistep thermal process assisting efficient isolation of islets from the FF pancreas and, subsequently, their dissociation into single islet cells. Hydroxylamine-based chemical decrosslinking helped restore intact peptide signals from individual isolated cells. Subsequently, an acetone/glycerol-assisted cell dispersion was optimized for spatially resolved cell deposition onto glass slides, while a glycerol solution maintained the hydrated state of the cells. This sample preparation procedure allowed peptide profiling in FF single cells by fluorescence-guided matrix-assisted laser desorption ionization MS. Here, 2594 single islet cells were analyzed and 28 peptides were detected, including insulin C-peptides and glucagon. T-distributed stochastic neighbor embedding (t-SNE) data visualization demonstrated that cells cluster based on cell-specific pancreatic peptide hormones. This workflow expands the sample availability for single-cell MS characterization to a wide range of formaldehyde-treated tissue specimens stored in biobanks.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Shapiro E; Biezuner T; Linnarsson S, Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature Reviews Genetics 2013, 14 9, 618–630. - PubMed
-
- Zhang L; Vertes A, Single-Cell Mass Spectrometry Approaches to Explore Cellular Heterogeneity. Angewandte Chemie International Edition 2018, 57 17, 4466–4477. - PubMed
-
- Wu Y; Chang Y; Shao Y; Guo G; Liu Z; Wang X, Controllable Fabrication of Small-Size Holding Pipets for the Nondestructive Manipulation of Suspended Living Single Cells. Analytical Chemistry 2022, 94 12, 4924–4929. - PubMed
-
- Lombard-Banek C; Li J; Portero EP; Onjiko RM; Singer CD; Plotnick DO; Al Shabeeb RQ; Nemes P, In Vivo Subcellular Mass Spectrometry Enables Proteo-Metabolomic Single-Cell Systems Biology in a Chordate Embryo Developing to a Normally Behaving Tadpole (X. laevis)**. 2021, 60 23, 12852–12858. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

