Cauliflower mosaic virus protein P6 is a multivalent node for RNA granule proteins and interferes with stress granule responses during plant infection
- PMID: 37040611
- PMCID: PMC10473198
- DOI: 10.1093/plcell/koad101
Cauliflower mosaic virus protein P6 is a multivalent node for RNA granule proteins and interferes with stress granule responses during plant infection
Abstract
Biomolecular condensation is a multipurpose cellular process that viruses use ubiquitously during their multiplication. Cauliflower mosaic virus replication complexes are condensates that differ from those of most viruses, as they are nonmembranous assemblies that consist of RNA and protein, mainly the viral protein P6. Although these viral factories (VFs) were described half a century ago, with many observations that followed since, functional details of the condensation process and the properties and relevance of VFs have remained enigmatic. Here, we studied these issues in Arabidopsis thaliana and Nicotiana benthamiana. We observed a large dynamic mobility range of host proteins within VFs, while the viral matrix protein P6 is immobile, as it represents the central node of these condensates. We identified the stress granule (SG) nucleating factors G3BP7 and UBP1 family members as components of VFs. Similarly, as SG components localize to VFs during infection, ectopic P6 localizes to SGs and reduces their assembly after stress. Intriguingly, it appears that soluble rather than condensed P6 suppresses SG formation and mediates other essential P6 functions, suggesting that the increased condensation over the infection time-course may accompany a progressive shift in selected P6 functions. Together, this study highlights VFs as dynamic condensates and P6 as a complex modulator of SG responses.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures

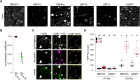
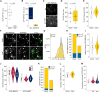
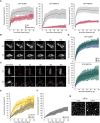
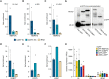

Similar articles
-
Cauliflower mosaic virus P6 Dysfunctions Histone Deacetylase HD2C to Promote Virus Infection.Cells. 2021 Sep 1;10(9):2278. doi: 10.3390/cells10092278. Cells. 2021. PMID: 34571927 Free PMC article.
-
The open reading frame VI product of Cauliflower mosaic virus is a nucleocytoplasmic protein: its N terminus mediates its nuclear export and formation of electron-dense viroplasms.Plant Cell. 2005 Mar;17(3):927-43. doi: 10.1105/tpc.104.029017. Plant Cell. 2005. PMID: 15746075 Free PMC article.
-
Association of the P6 protein of Cauliflower mosaic virus with plasmodesmata and plasmodesmal proteins.Plant Physiol. 2014 Nov;166(3):1345-58. doi: 10.1104/pp.114.249250. Epub 2014 Sep 19. Plant Physiol. 2014. PMID: 25239023 Free PMC article.
-
Joining the Crowd: Integrating Plant Virus Proteins into the Larger World of Pathogen Effectors.Annu Rev Phytopathol. 2018 Aug 25;56:89-110. doi: 10.1146/annurev-phyto-080417-050151. Epub 2018 May 31. Annu Rev Phytopathol. 2018. PMID: 29852091 Review.
-
A model for intracellular movement of Cauliflower mosaic virus: the concept of the mobile virion factory.J Exp Bot. 2016 Mar;67(7):2039-48. doi: 10.1093/jxb/erv520. Epub 2015 Dec 18. J Exp Bot. 2016. PMID: 26687180 Review.
Cited by
-
Viral proteins suppress rice defenses by boosting OsTSN1 RNA decay via phase separation and multimerization.Nat Commun. 2025 Aug 12;16(1):7481. doi: 10.1038/s41467-025-62395-5. Nat Commun. 2025. PMID: 40796552 Free PMC article.
-
A prion-like domain is required for phase separation and chloroplast RNA processing during cold acclimation in Arabidopsis.Plant Cell. 2024 Jul 31;36(8):2851-2872. doi: 10.1093/plcell/koae145. Plant Cell. 2024. PMID: 38723165 Free PMC article.
-
Focus on biomolecular condensates.Plant Cell. 2023 Sep 1;35(9):3155-3157. doi: 10.1093/plcell/koad182. Plant Cell. 2023. PMID: 37352160 Free PMC article. No abstract available.
-
Phase separation-based visualization of protein-protein interactions and kinase activities in plants.Plant Cell. 2023 Sep 1;35(9):3280-3302. doi: 10.1093/plcell/koad188. Plant Cell. 2023. PMID: 37378595 Free PMC article.
-
Biomolecular condensates in plant immunity.Cell Host Microbe. 2025 Aug 13;33(8):1276-1290. doi: 10.1016/j.chom.2025.06.014. Cell Host Microbe. 2025. PMID: 40812178 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

