Comparative epidemic expansion of SARS-CoV-2 variants Delta and Omicron in the Brazilian State of Amazonas
- PMID: 37041143
- PMCID: PMC10089528
- DOI: 10.1038/s41467-023-37541-6
Comparative epidemic expansion of SARS-CoV-2 variants Delta and Omicron in the Brazilian State of Amazonas
Abstract
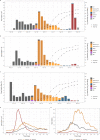
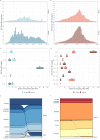
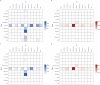
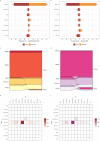
The SARS-CoV-2 variants of concern (VOCs) Delta and Omicron spread globally during mid and late 2021, respectively. In this study, we compare the dissemination dynamics of these VOCs in the Amazonas state, one of Brazil's most heavily affected regions. We sequenced the virus genome from 4128 patients collected in Amazonas between July 1st, 2021, and January 31st, 2022, and investigated the viral dynamics using a phylodynamic approach. The VOCs Delta and Omicron BA.1 displayed similar patterns of phylogeographic spread but different epidemic dynamics. The replacement of Gamma by Delta was gradual and occurred without an upsurge of COVID-19 cases, while the rise of Omicron BA.1 was extremely fast and fueled a sharp increase in cases. Thus, the dissemination dynamics and population-level impact of new SARS-CoV-2 variants introduced in the Amazonian population after mid-2021, a setting with high levels of acquired immunity, greatly vary according to their viral phenotype.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

