USP1 modulates hepatocellular carcinoma progression via the Hippo/TAZ axis
- PMID: 37041150
- PMCID: PMC10090121
- DOI: 10.1038/s41419-023-05777-1
USP1 modulates hepatocellular carcinoma progression via the Hippo/TAZ axis
Abstract
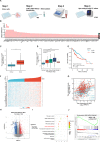
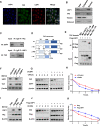
Hepatocellular carcinoma (HCC) is one of the most lethal malignancies worldwide. The Hippo signaling pathway has emerged as a significant suppressive pathway for hepatocellular carcinogenesis. The core components of the Hippo pathway constitute a kinase cascade, which inhibits the functional activation of YAP/TAZ. Interestingly, the overactivation of YAP/TAZ is commonly observed in hepatocellular carcinoma, although the inhibitory kinase cascade of the Hippo pathway is still functional. Recent studies have indicated that the ubiquitin‒proteasome system also plays important roles in modulating Hippo signaling activity. Our DUB (deubiquitinase) siRNA screen showed that USP1 is a critical regulator of Hippo signaling activity. Analysis of TCGA data demonstrated that USP1 expression is elevated in HCC and associated with poor survival in HCC patients. RNA sequencing analysis revealed that USP1 depletion affects Hippo signaling activity in HCC cell lines. Mechanistic assays revealed that USP1 is required for Hippo/TAZ axis activity and HCC progression. USP1 interacted with the WW domain of TAZ, which subsequently enhanced TAZ stability by suppressing K11-linked polyubiquitination of TAZ. Our study identifies a novel mechanism linking USP1 and TAZ in regulating the Hippo pathway and one possible therapeutic target for HCC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

