FADS1-arachidonic acid axis enhances arachidonic acid metabolism by altering intestinal microecology in colorectal cancer
- PMID: 37041160
- PMCID: PMC10090135
- DOI: 10.1038/s41467-023-37590-x
FADS1-arachidonic acid axis enhances arachidonic acid metabolism by altering intestinal microecology in colorectal cancer
Abstract
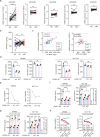
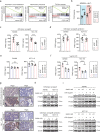
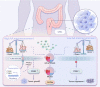
Colonocyte metabolism shapes the microbiome. Metabolites are the main mediators of information exchange between intestine and microbial communities. Arachidonic acid (AA) is an essential polyunsaturated fatty acid and its role in colorectal cancer (CRC) remains unexplored. In this study, we show that AA feeding promotes tumor growth in AOM/DSS and intestinal specific Apc-/- mice via modulating the intestinal microecology of increased gram-negative bacteria. Delta-5 desaturase (FADS1), a rate-limiting enzyme, is upregulated in CRC and effectively mediates AA synthesis. Functionally, FADS1 regulates CRC tumor growth via high AA microenvironment-induced enriched gram-negative microbes. Elimination of gram-negative microbe abolishes FADS1 effect. Mechanistically, gram-negative microbes activate TLR4/MYD88 pathway in CRC cells that contributes FADS1-AA axis to metabolize to prostaglandin E2 (PGE2). Cumulatively, we report a potential cancer-promoting mechanism of FADS1-AA axis in CRC that converts raising synthesized AA to PGE2 via modulating the intestinal microecology of gram-negative.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

