A bacterial autotransporter impairs innate immune responses by targeting the transcription factor TFE3
- PMID: 37041208
- PMCID: PMC10090168
- DOI: 10.1038/s41467-023-37812-2
A bacterial autotransporter impairs innate immune responses by targeting the transcription factor TFE3
Abstract
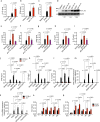
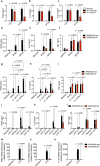
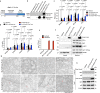
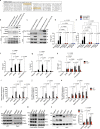
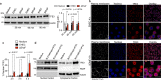
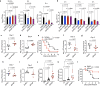
Type I interferons (IFNs) are consequential cytokines in antibacterial defense. Whether and how bacterial pathogens inhibit innate immune receptor-driven type I IFN expression remains mostly unknown. By screening a library of enterohemorrhagic Escherichia coli (EHEC) mutants, we uncovered EhaF, an uncharacterized protein, as an inhibitor of innate immune responses including IFNs. Further analyses identified EhaF as a secreted autotransporter-a type of bacterial secretion system with no known innate immune-modulatory function-that translocates into host cell cytosol and inhibit IFN response to EHEC. Mechanistically, EhaF interacts with and inhibits the MiT/TFE family transcription factor TFE3 resulting in impaired TANK phosphorylation and consequently, reduced IRF3 activation and type I IFN expression. Notably, EhaF-mediated innate immune suppression promotes EHEC colonization and pathogenesis in vivo. Overall, this study has uncovered a previously unknown autotransporter-based bacterial strategy that targets a specific transcription factor to subvert innate host defense.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
eIF4A3 Promotes RNA Viruses' Replication by Inhibiting Innate Immune Responses.J Virol. 2022 Nov 23;96(22):e0151322. doi: 10.1128/jvi.01513-22. Epub 2022 Oct 31. J Virol. 2022. PMID: 36314820 Free PMC article.
-
Heat Shock-Binding Protein 21 Regulates the Innate Immune Response to Viral Infection.J Virol. 2022 Apr 13;96(7):e0000122. doi: 10.1128/jvi.00001-22. Epub 2022 Mar 7. J Virol. 2022. PMID: 35254105 Free PMC article.
-
EspZ of enteropathogenic and enterohemorrhagic Escherichia coli regulates type III secretion system protein translocation.mBio. 2012 Oct 2;3(5):e00317-12. doi: 10.1128/mBio.00317-12. Print 2012. mBio. 2012. PMID: 23033475 Free PMC article.
-
Unraveling enterohemorrhagic Escherichia coli infection: the promising role of dietary compounds and probiotics in bacterial elimination and host innate immunity boosting.Crit Rev Food Sci Nutr. 2023;63(11):1551-1563. doi: 10.1080/10408398.2021.1965538. Epub 2021 Aug 18. Crit Rev Food Sci Nutr. 2023. PMID: 34404306 Review.
-
Key Roles of MiT Transcription Factors in Innate Immunity and Inflammation.Trends Immunol. 2020 Feb;41(2):157-171. doi: 10.1016/j.it.2019.12.003. Epub 2020 Jan 17. Trends Immunol. 2020. PMID: 31959514 Free PMC article. Review.
Cited by
-
Host-Pathogen Interactions during Shiga Toxin-Producing Escherichia coli Adherence and Colonization in the Bovine Gut: A Comprehensive Review.Microorganisms. 2024 Oct 3;12(10):2009. doi: 10.3390/microorganisms12102009. Microorganisms. 2024. PMID: 39458318 Free PMC article. Review.
-
A bacterial toxin co-opts caspase-3 to disable active gasdermin D and limit macrophage pyroptosis.Cell Rep. 2024 Apr 23;43(4):114004. doi: 10.1016/j.celrep.2024.114004. Epub 2024 Mar 22. Cell Rep. 2024. PMID: 38522070 Free PMC article.
References
-
- Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

