Single-cell RNA sequencing deciphers the mechanism of sepsis-induced liver injury and the therapeutic effects of artesunate
- PMID: 37041228
- PMCID: PMC10462669
- DOI: 10.1038/s41401-023-01065-y
Single-cell RNA sequencing deciphers the mechanism of sepsis-induced liver injury and the therapeutic effects of artesunate
Abstract
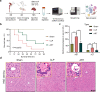
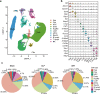
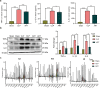
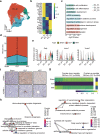
Liver, as an immune and detoxification organ, represents an important line of defense against bacteria and infection and a vulnerable organ that is easily injured during sepsis. Artesunate (ART) is an anti-malaria agent, that also exhibits broad pharmacological activities including anti-inflammatory, immune-regulation and liver protection. In this study, we investigated the cellular responses in liver to sepsis infection and ART hepatic-protective mechanisms against sepsis. Cecal ligation and puncture (CLP)-induced sepsis model was established in mice. The mice were administered ART (10 mg/kg, i.p.) at 4 h, and sacrificed at 12 h after the surgery. Liver samples were collected for preparing single-cell RNA transcriptome sequencing (scRNA-seq). The scRNA-seq analysis revealed that sepsis-induced a dramatic reduction of hepatic endothelial cells, especially the subtypes characterized with proliferation and differentiation. Macrophages were recruited during sepsis and released inflammatory cytokines (Tnf, Il1b, Il6), chemokines (Ccl6, Cd14), and transcription factor (Nfkb1), resulting in liver inflammatory responses. Massive apoptosis of lymphocytes and abnormal recruitment of neutrophils caused immune dysfunction. ART treatment significantly improved the survival of CLP mice within 96 h, and partially relieved or reversed the above-mentioned pathological features, mitigating the impact of sepsis on liver injury, inflammation, and dysfunction. This study provides comprehensive fundamental proof for the liver protective efficacy of ART against sepsis infection, which would potentially contribute to its clinical translation for sepsis therapy. Single cell transcriptome reveals the changes of various hepatocyte subtypes of CLP-induced liver injury and the potential pharmacological effects of artesunate on sepsis.
Keywords: sepsis; scRNA-seq; artesunate; liver; hepatic endothelial cells; macrophages; lymphocytes; neutrophils.
© 2023. The Author(s), under exclusive licence to Shanghai Institute of Materia Medica, Chinese Academy of Sciences and Chinese Pharmacological Society.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

