Improving the response to oxaliplatin by targeting chemotherapy-induced CLDN1 in resistant metastatic colorectal cancer cells
- PMID: 37041570
- PMCID: PMC10091849
- DOI: 10.1186/s13578-023-01015-5
Improving the response to oxaliplatin by targeting chemotherapy-induced CLDN1 in resistant metastatic colorectal cancer cells
Abstract
Background: Tumor resistance is a frequent cause of therapy failure and remains a major challenge for the long-term management of colorectal cancer (CRC). The aim of this study was to determine the implication of the tight junctional protein claudin 1 (CLDN1) in the acquired resistance to chemotherapy.
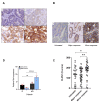
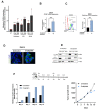
Methods: Immunohistochemistry was used to determine CLDN1 expression in post-chemotherapy liver metastases from 58 CRC patients. The effects of oxaliplatin on membrane CLDN1 expression were evaluated by flow cytometry, immunofluorescence and western blotting experiments in vitro and in vivo. Phosphoproteome analyses, proximity ligation and luciferase reporter assays were used to unravel the mechanism of CLDN1 induction. RNAseq experiments were performed on oxaliplatin-resistant cell lines to investigate the role of CLDN1 in chemoresistance. The "one-two punch" sequential combination of oxaliplatin followed by an anti-CLDN1 antibody-drug conjugate (ADC) was tested in both CRC cell lines and murine models.
Results: We found a significant correlation between CLDN1 expression level and histologic response to chemotherapy, CLDN1 expression being the highest in resistant metastatic residual cells of patients showing minor responses. Moreover, in both murine xenograft model and CRC cell lines, CLDN1 expression was upregulated after exposure to conventional chemotherapies used in CRC treatment. CLDN1 overexpression was, at least in part, functionally related to the activation of the MAPKp38/GSK3β/Wnt/β-catenin pathway. Overexpression of CLDN1 was also observed in oxaliplatin-resistant CRC cell lines and was associated with resistance to apoptosis, suggesting an anti-apoptotic role for CLDN1. Finally, we demonstrated that the sequential treatment with oxaliplatin followed by an anti-CLDN1 ADC displayed a synergistic effect in vitro and in in vivo.
Conclusion: Our study identifies CLDN1 as a new biomarker of acquired resistance to chemotherapy in CRC patients and suggests that a "one-two punch" approach targeting chemotherapy-induced CLDN1 expression may represent a therapeutic opportunity to circumvent resistance and to improve the outcome of patients with advanced CRC.
Keywords: ADC; Biomarker; CLDN1; Chemotherapy; Colorectal cancer; Resistance; “One-two punch”.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Yanagisawa Y, Maruta F, Iinuma N, Ishizone S, Koide N, Nakayama J, et al. Modified Irinotecan/5FU/Leucovorin therapy in advanced colorectal cancer and predicting therapeutic efficacy by expression of tumor-related enzymes. Scand J Gastroenterol. 2007;42:477–84. doi: 10.1080/00365520600994418. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

