The mycobiome of a successful crayfish invader and its changes along the environmental gradient
- PMID: 37041598
- PMCID: PMC10088235
- DOI: 10.1186/s42523-023-00245-9
The mycobiome of a successful crayfish invader and its changes along the environmental gradient
Abstract
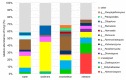
Background: The microbiome plays an important role in biological invasions, since it affects various interactions between host and environment. However, most studies focus on the bacteriome, insufficiently addressing other components of the microbiome such as the mycobiome. Microbial fungi are among the most damaging pathogens in freshwater crayfish populations, colonizing and infecting both native and invasive crayfish species. Invading crayfish may transmit novel fungal species to native populations, but also, dispersal process and characteristics of the novel environment may affect the invaders' mycobiome composition, directly and indirectly affecting their fitness and invasion success. This study analyzes the mycobiome of a successful invader in Europe, the signal crayfish, using the ITS rRNA amplicon sequencing approach. We explored the mycobiomes of crayfish samples (exoskeletal biofilm, hemolymph, hepatopancreas, intestine), compared them to environmental samples (water, sediment), and examined the differences in fungal diversity and abundance between upstream and downstream segments of the signal crayfish invasion range in the Korana River, Croatia.
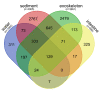
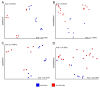
Results: A low number of ASVs (indicating low abundance and/or diversity of fungal taxa) was obtained in hemolymph and hepatopancreas samples. Thus, only exoskeleton, intestine, sediment and water samples were analyzed further. Significant differences were recorded between their mycobiomes, confirming their uniqueness. Generally, environmental mycobiomes showed higher diversity than crayfish-associated mycobiomes. The intestinal mycobiome showed significantly lower richness compared to other mycobiomes. Significant differences in the diversity of sediment and exoskeletal mycobiomes were recorded between different river segments (but not for water and intestinal mycobiomes). Together with the high observed portion of shared ASVs between sediment and exoskeleton, this indicates that the environment (i.e. sediment mycobiome) at least partly shapes the exoskeletal mycobiome of crayfish.
Conclusion: This study presents the first data on crayfish-associated fungal communities across different tissues, which is valuable given the lack of studies on the crayfish mycobiome. We demonstrate significant differences in the crayfish exoskeletal mycobiome along the invasion range, suggesting that different local environmental conditions may shape the exoskeletal mycobiome during range expansion, while the mycobiome of the internal organ (intestine) remained more stable. Our results provide a basis for assessing how the mycobiome contributes to the overall health of the signal crayfish and its further invasion success.
Keywords: Fungi; ITS rRNA gene; Invasive species; Microbiome; Pacifastacus leniusculus.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Microbiome of the Successful Freshwater Invader, the Signal Crayfish, and Its Changes along the Invasion Range.Microbiol Spectr. 2021 Oct 31;9(2):e0038921. doi: 10.1128/Spectrum.00389-21. Epub 2021 Sep 8. Microbiol Spectr. 2021. PMID: 34494878 Free PMC article.
-
Host-related traits influence the microbial diversity of the invasive signal crayfish Pacifastacus leniusculus.J Invertebr Pathol. 2024 Feb;202:108039. doi: 10.1016/j.jip.2023.108039. Epub 2023 Dec 12. J Invertebr Pathol. 2024. PMID: 38097037
-
Comparison of exoskeleton microbial communities of co-occurring native and invasive crayfish species.J Invertebr Pathol. 2023 Nov;201:107996. doi: 10.1016/j.jip.2023.107996. Epub 2023 Sep 30. J Invertebr Pathol. 2023. PMID: 37783231
-
Fungi of the human eye: Culture to mycobiome.Exp Eye Res. 2022 Apr;217:108968. doi: 10.1016/j.exer.2022.108968. Epub 2022 Feb 1. Exp Eye Res. 2022. PMID: 35120870 Review.
-
The human lung and Aspergillus: You are what you breathe in?Med Mycol. 2019 Apr 1;57(Supplement_2):S145-S154. doi: 10.1093/mmy/myy149. Med Mycol. 2019. PMID: 30816978 Free PMC article. Review.
Cited by
-
Industrial diet intervention modulates the interplay between gut microbiota and host in semi-stray dogs.Anim Microbiome. 2024 Nov 21;6(1):69. doi: 10.1186/s42523-024-00357-w. Anim Microbiome. 2024. PMID: 39574203 Free PMC article.
References
-
- Pyšek P, Richardson DM. Invasive species, Environmental Change and Management, and Health. Annu Rev Environ Resour. 2010;35:25–55. doi: 10.1146/annurev-environ-033009-095548. - DOI
-
- Ooue K, Ishiyama N, Ichimura M, Nakamura F. Environmental factors affecting the invasion success and morphological responses of a globally introduced crayfish in floodplain waterbodies. Biol Invasions. 2019;21:2639–52. doi: 10.1007/s10530-019-02008-7. - DOI
-
- Ni M, Deane DC, Li S, Wu Y, Sui X, Xu H, et al. Invasion success and impacts depend on different characteristics in non-native plants. Divers Distrib. 2021;27:1194–207. doi: 10.1111/ddi.13267. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
