Characterizing nrDNA ITS1, 5.8S and ITS2 secondary structures and their phylogenetic utility in the legume tribe Hedysareae with special reference to Hedysarum
- PMID: 37043495
- PMCID: PMC10096232
- DOI: 10.1371/journal.pone.0283847
Characterizing nrDNA ITS1, 5.8S and ITS2 secondary structures and their phylogenetic utility in the legume tribe Hedysareae with special reference to Hedysarum
Abstract
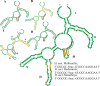
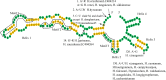
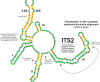
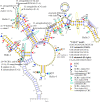
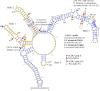
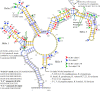
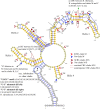
This is the first study to systematically evaluate rRNA secondary structures of Hedysareae with an emphasis on Hedysarum. ITS2 and 5.8S regions of the genus shared a common secondary structure with a four-fingered central loop, whereas ITS1 possessed five distinct structures. The secondary structural features of the two regions provided advantageous data for clades, species groups, and closely related species. Hemi-CBCs were mostly observed in the reconstruction of species groups, and Nsts, mostly between closely related species. The investigations showed that ITS1 varied more than ITS2 in length, GC content, and most of the diversity indices within the tribe. Maximum likelihood analyses of the synchronized sequence-structure tree of ITS1 were performed. The accuracy and phylogenetic signals of ITS1 were higher than ITS2. The similar GC content, and no CBC, in both spacers, fortified the close relationship of CEGO and H. sections Stracheya and Hedysarum clades in the synchronized sequence-structure tree topology of ITS1. In both regions, no inter-generic CBCs were detected inside the CEGO clade and the inter-sectional level of Hedysarum. But, in the ITS2 region, a CBC was detected between H. section Multicaulia, and Taverniera versus H. sections Hedysarum, and Stracheya. The lowest inter-sectional genetic distance and structural features were found between H. sect. Hedysarum and H. sect. Stracheya clades in the ITS2 region.
Copyright: © 2023 Nafisi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Hedysarum L. (Fabaceae: Hedysareae) Is Not Monophyletic - Evidence from Phylogenetic Analyses Based on Five Nuclear and Five Plastid Sequences.PLoS One. 2017 Jan 25;12(1):e0170596. doi: 10.1371/journal.pone.0170596. eCollection 2017. PLoS One. 2017. PMID: 28122062 Free PMC article.
-
Species of the Sections Hedysarum and Multicaulia of the Genus Hedysarum (Fabaceae): Taxonomy, Distribution, Chromosomes, Genomes, and Phylogeny.Int J Mol Sci. 2024 Aug 3;25(15):8489. doi: 10.3390/ijms25158489. Int J Mol Sci. 2024. PMID: 39126057 Free PMC article. Review.
-
A close-up view on ITS2 evolution and speciation - a case study in the Ulvophyceae (Chlorophyta, Viridiplantae).BMC Evol Biol. 2011 Sep 20;11:262. doi: 10.1186/1471-2148-11-262. BMC Evol Biol. 2011. PMID: 21933414 Free PMC article.
-
First comparative analysis of complete chloroplast genomes among six Hedysarum (Fabaceae) species.Front Plant Sci. 2023 Aug 18;14:1211247. doi: 10.3389/fpls.2023.1211247. eCollection 2023. Front Plant Sci. 2023. PMID: 37662153 Free PMC article.
-
Phylogenetic Utility of rRNA ITS2 Sequence-Structure under Functional Constraint.Int J Mol Sci. 2020 Sep 3;21(17):6395. doi: 10.3390/ijms21176395. Int J Mol Sci. 2020. PMID: 32899108 Free PMC article. Review.
Cited by
-
DNA barcoding, micromorphology and metabolic traits of selected Ficus L. (Moraceae) species from Egypt.BMC Plant Biol. 2024 Nov 13;24(1):1067. doi: 10.1186/s12870-024-05683-4. BMC Plant Biol. 2024. PMID: 39538137 Free PMC article.
References
-
- Duan L, Wen J, Yang X, Liu PL, Arslan E, Ertuğrul K, et al.. Phylogeny of Hedysarum and tribe Hedysareae (Leguminosae: Papilionoideae) inferred from sequence data of ITS, matK, trnL-F and psbA-trnH. Taxon. 2015;64: 49–64. doi: 10.12705/641.26 - DOI
-
- Nafisi H, Kazempour-Osaloo Sh, Kaveh A, Mahmoodi MA. Taxonomic Revision of the Genus Hedysarum L. (Fabaceae-Hedysareae) in Iran. Phytotaxa. 2021;511: 079–110. doi: 10.11646/phytotaxa.511.2.1 - DOI
-
- Nafisi H, Kazempour-Osaloo S, Mozaffarian V, Schneeweiss M. Molecular phylogeny and divergence times of the genus Hedysarum (Fabaceae) with special reference to section Multicaulia in Southwest Asia. Plant Syst Evol. 2019;305: 1001–1017. Available from: doi: 10.1007/s00606-019-01620-3 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

