In SARS-CoV-2 delta variants, Spike-P681R and D950N promote membrane fusion, Spike-P681R enhances spike cleavage, but neither substitution affects pathogenicity in hamsters
- PMID: 37043872
- PMCID: PMC10083686
- DOI: 10.1016/j.ebiom.2023.104561
In SARS-CoV-2 delta variants, Spike-P681R and D950N promote membrane fusion, Spike-P681R enhances spike cleavage, but neither substitution affects pathogenicity in hamsters
Abstract
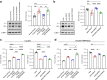
Background: The SARS-CoV-2 delta (B.1.617.2 lineage) variant was first identified at the end of 2020 and possessed two unique amino acid substitutions in its spike protein: S-P681R, at the S1/S2 cleavage site, and S-D950N, in the HR1 of the S2 subunit. However, the roles of these substitutions in virus phenotypes have not been fully characterized.
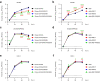
Methods: We used reverse genetics to generate Wuhan-D614G viruses with these substitutions and delta viruses lacking these substitutions and explored how these changes affected their viral characteristics in vitro and in vivo.
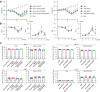
Findings: S-P681R enhanced spike cleavage and membrane fusion, whereas S-D950N slightly promoted membrane fusion. Although S-681R reduced the virus replicative ability especially in VeroE6 cells, neither substitution affected virus replication in Calu-3 cells and hamsters. The pathogenicity of all recombinant viruses tested in hamsters was slightly but not significantly affected.
Interpretation: Our observations suggest that the S-P681R and S-D950N substitutions alone do not increase virus pathogenicity, despite of their enhancement of spike cleavage or fusogenicity.
Funding: A full list of funding bodies that contributed to this study can be found under Acknowledgments.
Keywords: COVID-19; Hamster; Reverse genetics; SARS-CoV-2.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests Y.K. has received collaborative research funds from the following companies. For antiviral studies; FUJIFILM Toyama Chemical Co. LTD (various viruses), Shionogi &; Co. LTD (influenza viruses). For vaccine and other studies; Daiichi Sankyo Pharmaceutical, Otsuka Pharmaceutical, KM Biologics, Kyoritsu Seiyaku, Fuji Rebio, Tauns Laboratories, Inc., Matsubara Co. LTD. As the Flagship Center of the Japan Initiative for World-leading Vaccine Research and Development Centers, we plan to initiate collaborative research related to vaccine development with the following companies: Shionogi &Co. LTD, Daiichi Sankyo Pharmaceutical, KM Biologics, Cytiva, Sysmex Corporation, NEC.
Figures

Similar articles
-
Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation.Nature. 2022 Feb;602(7896):300-306. doi: 10.1038/s41586-021-04266-9. Epub 2021 Nov 25. Nature. 2022. PMID: 34823256 Free PMC article.
-
Spike Protein Cleavage-Activation in the Context of the SARS-CoV-2 P681R Mutation: an Analysis from Its First Appearance in Lineage A.23.1 Identified in Uganda.Microbiol Spectr. 2022 Aug 31;10(4):e0151422. doi: 10.1128/spectrum.01514-22. Epub 2022 Jun 29. Microbiol Spectr. 2022. PMID: 35766497 Free PMC article.
-
Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant.Cell Rep. 2022 May 17;39(7):110829. doi: 10.1016/j.celrep.2022.110829. Epub 2022 Apr 29. Cell Rep. 2022. PMID: 35550680 Free PMC article.
-
Intrinsic D614G and P681R/H mutations in SARS-CoV-2 VoCs Alpha, Delta, Omicron and viruses with D614G plus key signature mutations in spike protein alters fusogenicity and infectivity.Med Microbiol Immunol. 2023 Feb;212(1):103-122. doi: 10.1007/s00430-022-00760-7. Epub 2022 Dec 30. Med Microbiol Immunol. 2023. PMID: 36583790 Free PMC article.
-
Spike protein cleavage-activation mediated by the SARS-CoV-2 P681R mutation: a case-study from its first appearance in variant of interest (VOI) A.23.1 identified in Uganda.bioRxiv [Preprint]. 2022 Mar 28:2021.06.30.450632. doi: 10.1101/2021.06.30.450632. bioRxiv. 2022. Update in: Microbiol Spectr. 2022 Aug 31;10(4):e0151422. doi: 10.1128/spectrum.01514-22. PMID: 34230931 Free PMC article. Updated. Preprint.
Cited by
-
Characterization of SARS-CoV-2 Variants in Military and Civilian Personnel of an Air Force Airport during Three Pandemic Waves in Italy.Microorganisms. 2023 Nov 5;11(11):2711. doi: 10.3390/microorganisms11112711. Microorganisms. 2023. PMID: 38004723 Free PMC article.
-
Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.EMBO J. 2024 Dec;43(24):6469-6495. doi: 10.1038/s44318-024-00303-1. Epub 2024 Nov 14. EMBO J. 2024. PMID: 39543395 Free PMC article.
-
Insights into Omicron's Low Fusogenicity through In Silico Molecular Studies on Spike-Furin Interactions.Bioinform Biol Insights. 2023 Jul 28;17:11779322231189371. doi: 10.1177/11779322231189371. eCollection 2023. Bioinform Biol Insights. 2023. PMID: 37529484 Free PMC article.
-
Pathogenesis and transmission of SARS-CoV-2 D614G, Alpha, Gamma, Delta, and Omicron variants in golden hamsters.Npj Viruses. 2025 Feb 24;3(1):15. doi: 10.1038/s44298-025-00092-2. Npj Viruses. 2025. PMID: 40295859 Free PMC article.
-
Profiling endogenous airway proteases and antiproteases and modeling proteolytic activation of Influenza HA using in vitro and ex vivo human airway surface liquid samples.PLoS One. 2024 Dec 31;19(12):e0306197. doi: 10.1371/journal.pone.0306197. eCollection 2024. PLoS One. 2024. PMID: 39739661 Free PMC article.
References
-
- Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. SARS-CoV-2 variants of concern are emerging in India. Nat Med. 2021;27:1131–1133. - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

