Pyrazinamide-resistant Tuberculosis Obscured From Common Targeted Molecular Diagnostics
- PMID: 37043916
- PMCID: PMC10317212
- DOI: 10.1016/j.drup.2023.100959
Pyrazinamide-resistant Tuberculosis Obscured From Common Targeted Molecular Diagnostics
Abstract
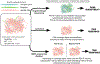
Here, we describe a clinical case of pyrazinamide-resistant (PZA-R) tuberculosis (TB) reported as PZA-susceptible (PZA-S) by common molecular diagnostics. Phenotypic susceptibility testing (pDST) indicated PZA-R TB. Targeted Sanger sequencing reported wild-type PncA, indicating PZA-S TB. Whole Genome Sequencing (WGS) by PacBio and IonTorrent both detected deletion of a large portion of pncA, indicating PZA-R. Importantly, both WGS methods showed deletion of part of the primer region targeted by Sanger sequencing. Repeating Sanger sequencing from a culture in presence of PZA returned no result, revealing that 1) two minority susceptible subpopulations had vanished, 2) the PZA-R majority subpopulation harboring the pncA deletion could not be amplified by Sanger primers, and was thus obscured by amplification process. This case demonstrates how a small susceptible subpopulation can entirely obscure majority resistant populations from targeted molecular diagnostics and falsely imply homogenous susceptibility, leading to incorrect diagnosis. To our knowledge, this is the first report of a minority susceptible subpopulation masking a majority resistant population, causing targeted molecular diagnostics to call false susceptibility. The consequence of such genomic events is not limited to PZA. This phenomenon can impact molecular diagnostics' sensitivity whenever the resistance-conferring mutation is not fully within primer-targeted regions. This can be caused by structural changes of genomic context with phenotypic consequence as we report here, or by uncommon mechanisms of resistance. Such false susceptibility calls promote suboptimal treatment and spread of strains that challenge targeted molecular diagnostics. This motivates development of molecular diagnostics unreliant on primer conservation, and impels frequent WGS surveillance for variants that evade prevailing molecular diagnostics.
Keywords: Heteroresistance; Molecular diagnostics; Pyrazinamide; Tuberculosis.
Copyright © 2023 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
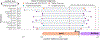
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

