Neonatal and adult cardiac fibroblasts exhibit inherent differences in cardiac regenerative capacity
- PMID: 37044217
- PMCID: PMC10193010
- DOI: 10.1016/j.jbc.2023.104694
Neonatal and adult cardiac fibroblasts exhibit inherent differences in cardiac regenerative capacity
Abstract
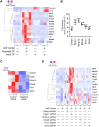
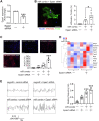
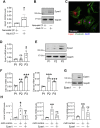
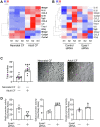
Directly reprogramming fibroblasts into cardiomyocytes improves cardiac function in the infarcted heart. However, the low efficacy of this approach hinders clinical applications. Unlike the adult mammalian heart, the neonatal heart has an intrinsic regenerative capacity. Consequently, we hypothesized that birth imposes fundamental changes in cardiac fibroblasts which limit their regenerative capabilities. In support, we found that reprogramming efficacy in vitro was markedly lower with fibroblasts derived from adult mice versus those derived from neonatal mice. Notably, fibroblasts derived from adult mice expressed significantly higher levels of pro-angiogenic genes. Moreover, under conditions that promote angiogenesis, only fibroblasts derived from adult mice differentiated into tube-like structures. Targeted knockdown screening studies suggested a possible role for the transcription factor Epas1. Epas1 expression was higher in fibroblasts derived from adult mice, and Epas1 knockdown improved reprogramming efficacy in cultured adult cardiac fibroblasts. Promoter activity assays indicated that Epas1 functions as both a transcription repressor and an activator, inhibiting cardiomyocyte genes while activating angiogenic genes. Finally, the addition of an Epas1 targeting siRNA to the reprogramming cocktail markedly improved reprogramming efficacy in vivo with both the number of reprogramming events and cardiac function being markedly improved. Collectively, our results highlight differences between neonatal and adult cardiac fibroblasts and the dual transcriptional activities of Epas1 related to reprogramming efficacy.
Keywords: aging; fibroblast; microRNA (miRNA); myocardial infarction; reprogramming.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest Conrad P. Hodgkinson and Victor J. Dzau are co-founders of Recardia Therapeutics. This company aims to translate miR combo to clinical applications. The remaining authors have no conflicts of interest.
Figures
References
-
- Jayawardena T., Mirotsou M., Dzau V.J. Direct reprogramming of cardiac fibroblasts to cardiomyocytes using microRNAs. Methods Mol. Biol. 2014;1150:263–272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

