Embryonic keratin19+ progenitors generate multiple functionally distinct progeny to maintain epithelial diversity in the adult thymus medulla
- PMID: 37045811
- PMCID: PMC10097809
- DOI: 10.1038/s41467-023-37589-4
Embryonic keratin19+ progenitors generate multiple functionally distinct progeny to maintain epithelial diversity in the adult thymus medulla
Abstract
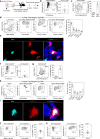
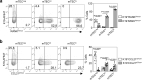
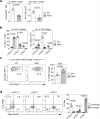
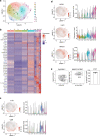
The thymus medulla is a key site for immunoregulation and tolerance, and its functional specialisation is achieved through the complexity of medullary thymic epithelial cells (mTEC). While the importance of the medulla for thymus function is clear, the production and maintenance of mTEC diversity remains poorly understood. Here, using ontogenetic and inducible fate-mapping approaches, we identify mTEC-restricted progenitors as a cytokeratin19+ (K19+) TEC subset that emerges in the embryonic thymus. Importantly, labelling of a single cohort of K19+ TEC during embryogenesis sustains the production of multiple mTEC subsets into adulthood, including CCL21+ mTEClo, Aire+ mTEChi and thymic tuft cells. We show K19+ progenitors arise prior to the acquisition of multiple mTEC-defining features including RANK and CCL21 and are generated independently of the key mTEC regulator, Relb. In conclusion, we identify and define a multipotent mTEC progenitor that emerges during embryogenesis to support mTEC diversity into adult life.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

