WHIM Syndrome-linked CXCR4 mutations drive osteoporosis
- PMID: 37045841
- PMCID: PMC10097661
- DOI: 10.1038/s41467-023-37791-4
WHIM Syndrome-linked CXCR4 mutations drive osteoporosis
Abstract
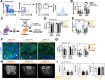
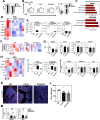
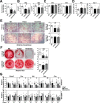
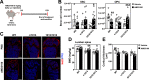
WHIM Syndrome is a rare immunodeficiency caused by gain-of-function CXCR4 mutations. Here we report a decrease in bone mineral density in 25% of WHIM patients and bone defects leading to osteoporosis in a WHIM mouse model. Imbalanced bone tissue is observed in mutant mice combining reduced osteoprogenitor cells and increased osteoclast numbers. Mechanistically, impaired CXCR4 desensitization disrupts cell cycle progression and osteogenic commitment of skeletal stromal/stem cells, while increasing their pro-osteoclastogenic capacities. Impaired osteogenic differentiation is evidenced in primary bone marrow stromal cells from WHIM patients. In mice, chronic treatment with the CXCR4 antagonist AMD3100 normalizes in vitro osteogenic fate of mutant skeletal stromal/stem cells and reverses in vivo the loss of skeletal cells, demonstrating that proper CXCR4 desensitization is required for the osteogenic specification of skeletal stromal/stem cells. Our study provides mechanistic insights into how CXCR4 signaling regulates the osteogenic fate of skeletal cells and the balance between bone formation and resorption.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

