Mutant and curli-producing E. coli enhance the disease phenotype in a hSOD1-G93A mouse model of ALS
- PMID: 37045868
- PMCID: PMC10097672
- DOI: 10.1038/s41598-023-32594-5
Mutant and curli-producing E. coli enhance the disease phenotype in a hSOD1-G93A mouse model of ALS
Abstract
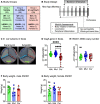
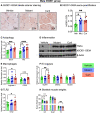
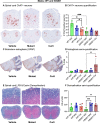
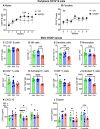
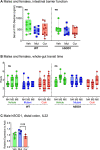
The gut microbiome is a potential non-genetic contributing factor for Amyotrophic Lateral Sclerosis. Differences in gut microbial communities have been detected between ALS subjects and healthy controls, including an increase in Escherichia coli in ALS subjects. E. coli and other gram-negative bacteria produce curli proteins, which are functional bacterial amyloids. We examined whether long-term curli overexposure in the gut can exacerbate the development and progression of ALS. We utilized the slow-developing hSOD1-G93A mouse model of ALS with their C57BL/6J WT littermate controls, including males and females, with a total of 91 animals. These mice were on a normal chow diet and fed curli-producing or curli-nonproducing (mutant) E. coli in applesauce (vehicle) 3 times/week, from 1 through 7 months of age. Male hSOD1 mice demonstrated gradual slowing in running speed month 4 onwards, while females exhibited no signs of locomotive impairment even at 7 months of age. Around the same time, male hSOD1 mice showed a gradual increase in frequency of peripheral CD19+ B cells. Among the male hSOD1 group, chronic gut exposure to curli-producing E. coli led to significant shifts in α- and β-diversities. Curli-exposed males showed suppression of immune responses in circulation, but an increase in markers of inflammation, autophagy and protein turnover in skeletal muscle. Some of these markers were also changed in mutant E. coli-exposed mice, including astrogliosis in the brainstem and demyelination in the lumbar spinal cord. Overall, chronic overexposure to a commensal bacteria like E. coli led to distant organ pathology in our model, without the presence of a leaky gut at 6 months. Mechanisms underlying gut-distant organ communication are of tremendous interest to all disciplines.
© 2023. The Author(s).
Conflict of interest statement
Robert P. Friedland is a stockholder in Axial Therapeutics. ZK, JDM, RAS, JM, SMS, ED, DAB, LJB, and LJS have no competing interests to declare.
Figures


References
-
- Kim S, Kwon SH, Kam TI, Panicker N, Karuppagounder SS, Lee S, Lee JH, Kim WR, Kook M, Foss CA, Shen C, Lee H, Kulkarni S, Pasricha PJ, Lee G, Pomper MG, Dawson VL, Dawson TM, Ko HS. Transneuronal propagation of pathologic alpha-synuclein from the gut to the brain models Parkinson’s disease. Neuron. 2019;103(627–641):e627. doi: 10.1016/j.neuron.2019.05.035. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

