Identification and panoramic analysis of drug response-related genes in triple negative breast cancer using as an example NVP-BEZ235
- PMID: 37045929
- PMCID: PMC10097725
- DOI: 10.1038/s41598-023-32757-4
Identification and panoramic analysis of drug response-related genes in triple negative breast cancer using as an example NVP-BEZ235
Abstract
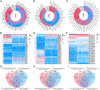
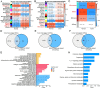
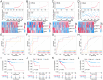
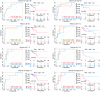
Taking NVP-BEZ235 (BEZ235) as an example to screen drug response-related genes (DRRGs) and explore their potential value in triple-negative breast cancer (TNBC). Through high-throughput technique, multidimensional transcriptome expression data (mRNA, miRNA and lncRNA) of BEZ235-treated and -untreated MDA-MB-468 cell lines were obtained. Combined with transcriptome data of the MDA-MB-468 cells and TCGA-TNBC tissues, differential gene expression analysis and WGCNA were performed to identify DRRGs associated with tumor trait by simulating the drug response microenvironment (DRM) of BEZ235-treated patients. Based on DRRGs, we constructed a ceRNA network and verified the expression levels of three key molecules by RT-qPCR, which not only demonstrated the successful construction of a BEZ235-treated cell line model but also explained the antitumor mechanism of BEZ235. Four molecular subtypes related to the DRM with survival difference were proposed using cluster analysis, namely glycolysis subtype, proliferation depression subtype, immune-suppressed subtype, and immune-activated subtype. A novel prognostic signature consisting of four DRRGs was established by Lasso-Cox analysis, which exhibited outstanding performance in predicting overall survival compared with several excellent reported signatures. The high- and low-risk groups were characterized by enrichment of metabolism-related pathways and immune-related pathways, respectively. Of note, the low-risk group had a better response to immune checkpoint blockade. Besides, pRRophetic analysis found that patients in the low-risk group were more sensitive to methotrexate and cisplation, whereas more resistant to BEZ235, docetaxel and paclitaxel. In conclusion, the DRRGs exemplified by BEZ235 are potential biomarkers for TNBC molecular typing, prognosis prediction and targeted therapy. The novel DRRGs-guided strategy for predicting the subtype, survival and therapy efficacy, might be also applied to more cancers and drugs other than TNBC and BEZ235.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

