Chromosomal fragile site breakage by EBV-encoded EBNA1 at clustered repeats
- PMID: 37046091
- PMCID: PMC10328181
- DOI: 10.1038/s41586-023-05923-x
Chromosomal fragile site breakage by EBV-encoded EBNA1 at clustered repeats
Abstract
Epstein-Barr virus (EBV) is an oncogenic herpesvirus associated with several cancers of lymphocytic and epithelial origin1-3. EBV encodes EBNA1, which binds to a cluster of 20 copies of an 18-base-pair palindromic sequence in the EBV genome4-6. EBNA1 also associates with host chromosomes at non-sequence-specific sites7, thereby enabling viral persistence. Here we show that the sequence-specific DNA-binding domain of EBNA1 binds to a cluster of tandemly repeated copies of an EBV-like, 18-base-pair imperfect palindromic sequence encompassing a region of about 21 kilobases at human chromosome 11q23. In situ visualization of the repetitive EBNA1-binding site reveals aberrant structures on mitotic chromosomes characteristic of inherently fragile DNA. We demonstrate that increasing levels of EBNA1 binding trigger dose-dependent breakage at 11q23, producing a fusogenic centromere-containing fragment and an acentric distal fragment, with both mis-segregated into micronuclei in the next cell cycles. In cells latently infected with EBV, elevating EBNA1 abundance by as little as twofold was sufficient to trigger breakage at 11q23. Examination of whole-genome sequencing of EBV-associated nasopharyngeal carcinomas revealed that structural variants are highly enriched on chromosome 11. Presence of EBV is also shown to be associated with an enrichment of chromosome 11 rearrangements across 2,439 tumours from 38 cancer types. Our results identify a previously unappreciated link between EBV and genomic instability, wherein EBNA1-induced breakage at 11q23 triggers acquisition of structural variations in chromosome 11.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Figures
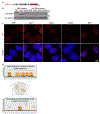


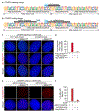
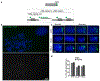
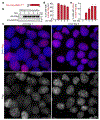

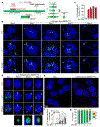
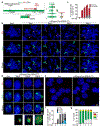
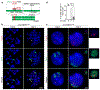
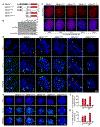
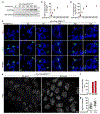
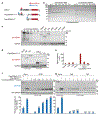
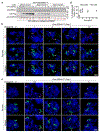

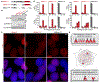
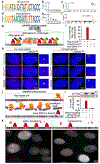

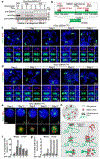
Comment in
-
Epstein-Barr virus is an agent of genomic instability.Nature. 2023 Apr;616(7957):441-442. doi: 10.1038/d41586-023-00936-y. Nature. 2023. PMID: 37045952 No abstract available.
References
-
- Pope JH, Horne MK & Scott W Transformation of foetal human keukocytes in vitro by filtrates of a human leukaemic cell line containing herpes-like virus. Int. J. Cancer 3, 857–866 (1968). - PubMed
-
- Hsu JL & Glaser SL Epstein-barr virus-associated malignancies: epidemiologic patterns and etiologic implications. Crit. Rev. Oncol. Hematol 34, 27–53 (2000). - PubMed
-
- Thorley-Lawson DA & Gross A Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med 350, 1328–1337 (2004). - PubMed
-
- Rawlins DR, Milman G, Hayward SD & Hayward GS Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42, 859–868 (1985). - PubMed
-
- Bochkarev A, et al. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell 83, 39–46 (1995). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

