Genome-wide analysis of circular RNA-mediated ceRNA regulation in porcine skeletal muscle development
- PMID: 37046223
- PMCID: PMC10099641
- DOI: 10.1186/s12864-023-09284-7
Genome-wide analysis of circular RNA-mediated ceRNA regulation in porcine skeletal muscle development
Abstract
Background: As a diverse and abundant class of endogenous RNAs, circular RNAs (circRNAs) participate in various biological processes including cell proliferation and apoptosis. Nevertheless, few researchers have investigated the role of circRNAs in muscle development in cultivated pigs.
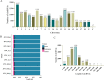
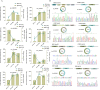
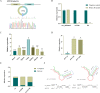
Results: In this study, we used RNA-seq to construct circRNA expression profiles in skeletal muscle of Jinfen White pigs at the age of 1, 90, and 180 days. Among the 16,990 identified circRNAs, 584 circRNAs were differentially expressed. Moreover, the enrichment analysis of DE circRNA host genes showed that they were mainly involved in muscle contraction, muscle organ development and muscle system processes, as well as AMPK and cAMP-related signal pathways. We also constructed a circRNA-miRNA-mRNA co-expression network to find key circRNAs which many involved in the regulation of porcine skeletal muscle development through the competitive endogenous RNA (ceRNA) mechanism. It is noteworthy that circ_0018595/miR-1343/PGM1 axis may play a regulatory role in the development of porcine skeletal muscle.
Conclusions: This study identified the circRNAs and present the circRNA expression profile in the development of pigs, revealed that DE circRNA host genes participate in different cell fates and enriched the porcine ceRNA network. Thus, this work will become a valuable resource for further in-depth study of the regulatory mechanism of circRNA in the development of porcine skeletal muscle.
Keywords: Pig; RNA-seq; Skeletal muscle; ceRNA; circRNA.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Comprehensive analysis of differentially expressed circRNAs and ceRNA regulatory network in porcine skeletal muscle.BMC Genomics. 2021 May 1;22(1):320. doi: 10.1186/s12864-021-07645-8. BMC Genomics. 2021. PMID: 33932987 Free PMC article.
-
Construction of circRNA-related ceRNA networks in longissimus dorsi muscle of Queshan Black and Large White pigs.Mol Genet Genomics. 2022 Jan;297(1):101-112. doi: 10.1007/s00438-021-01836-4. Epub 2021 Nov 18. Mol Genet Genomics. 2022. PMID: 34792645
-
Integrative analysis of circRNA, miRNA, and mRNA profiles to reveal ceRNA regulation in chicken muscle development from the embryonic to post-hatching periods.BMC Genomics. 2022 May 3;23(1):342. doi: 10.1186/s12864-022-08525-5. BMC Genomics. 2022. PMID: 35505302 Free PMC article.
-
Circular RNAs and host genes act synergistically in regulating cellular processes and functions in skeletal myogenesis.Gene. 2025 Mar 10;940:149189. doi: 10.1016/j.gene.2024.149189. Epub 2024 Dec 24. Gene. 2025. PMID: 39724991 Review.
-
RNA-Seq Revealed a Circular RNA-microRNA-mRNA Regulatory Network in Hantaan Virus Infection.Front Cell Infect Microbiol. 2020 Mar 13;10:97. doi: 10.3389/fcimb.2020.00097. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32232013 Free PMC article. Review.
Cited by
-
Differential gene expression drives muscle metabolic and structural differences in Liang Guang small spotted vs. large white pigs.Sci Rep. 2025 Aug 27;15(1):31564. doi: 10.1038/s41598-025-17179-8. Sci Rep. 2025. PMID: 40866463 Free PMC article.
-
Functional analysis of differentially expressed circular RNAs in sheep subcutaneous fat.BMC Genomics. 2023 Oct 5;24(1):591. doi: 10.1186/s12864-023-09401-6. BMC Genomics. 2023. PMID: 37798722 Free PMC article.
-
Characteristics of microRNAs in Skeletal Muscle of Intrauterine Growth-Restricted Pigs.Genes (Basel). 2023 Jun 28;14(7):1372. doi: 10.3390/genes14071372. Genes (Basel). 2023. PMID: 37510277 Free PMC article.
-
Combined analysis of lncRNA and mRNA emphasizes the potential role of tryptophan-mediated regulation of muscle development in weaned piglets by lncRNA.J Anim Sci. 2024 Jan 3;102:skae264. doi: 10.1093/jas/skae264. J Anim Sci. 2024. PMID: 39276131 Free PMC article.
-
CircRNA profiling of skeletal muscle satellite cells in goats reveals circTGFβ2 promotes myoblast differentiation.BMC Genomics. 2024 Nov 12;25(1):1075. doi: 10.1186/s12864-024-11008-4. BMC Genomics. 2024. PMID: 39533172 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

