Tracing Eastern Wolf Origins From Whole-Genome Data in Context of Extensive Hybridization
- PMID: 37046402
- PMCID: PMC10098045
- DOI: 10.1093/molbev/msad055
Tracing Eastern Wolf Origins From Whole-Genome Data in Context of Extensive Hybridization
Abstract
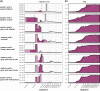
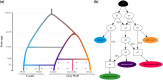
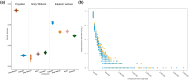
Southeastern Canada is inhabited by an amalgam of hybridizing wolf-like canids, raising fundamental questions regarding their taxonomy, origins, and timing of hybridization events. Eastern wolves (Canis lycaon), specifically, have been the subject of significant controversy, being viewed as either a distinct taxonomic entity of conservation concern or a recent hybrid of coyotes (C. latrans) and grey wolves (C. lupus). Mitochondrial DNA analyses show some evidence of eastern wolves being North American evolved canids. In contrast, nuclear genome studies indicate eastern wolves are best described as a hybrid entity, but with unclear timing of hybridization events. To test hypotheses related to these competing findings we sequenced whole genomes of 25 individuals, representative of extant Canadian wolf-like canid types of known origin and levels of contemporary hybridization. Here we present data describing eastern wolves as a distinct taxonomic entity that evolved separately from grey wolves for the past ∼67,000 years with an admixture event with coyotes ∼37,000 years ago. We show that Great Lakes wolves originated as a product of admixture between grey wolves and eastern wolves after the last glaciation (∼8,000 years ago) while eastern coyotes originated as a product of admixture between "western" coyotes and eastern wolves during the last century. Eastern wolf nuclear genomes appear shaped by historical and contemporary gene flow with grey wolves and coyotes, yet evolutionary uniqueness remains among eastern wolves currently inhabiting a restricted range in southeastern Canada.
Keywords: admixture; eastern coyote; eastern wolf; great lakes wolf; red wolf; whole genomes.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



Similar articles
-
Evolutionary legacy of the extirpated red wolf clings to life in gulf-coast canids.Mol Ecol. 2022 Nov;31(21):5419-5422. doi: 10.1111/mec.16725. Epub 2022 Oct 20. Mol Ecol. 2022. PMID: 36210646
-
Sympatric wolf and coyote populations of the western Great Lakes region are reproductively isolated.Mol Ecol. 2010 Oct;19(20):4428-40. doi: 10.1111/j.1365-294X.2010.04818.x. Epub 2010 Sep 13. Mol Ecol. 2010. PMID: 20854277
-
RAD sequencing and genomic simulations resolve hybrid origins within North American Canis.Biol Lett. 2015 Jul;11(7):20150303. doi: 10.1098/rsbl.2015.0303. Biol Lett. 2015. PMID: 26156129 Free PMC article.
-
Red Wolf (Canis rufus) Recovery: A Review with Suggestions for Future Research.Animals (Basel). 2013 Aug 13;3(3):722-44. doi: 10.3390/ani3030722. Animals (Basel). 2013. PMID: 26479530 Free PMC article. Review.
-
Evaluating the Taxonomic Status of the Mexican Gray Wolf and the Red Wolf.Washington (DC): National Academies Press (US); 2019 Mar 28. Washington (DC): National Academies Press (US); 2019 Mar 28. PMID: 31211533 Free Books & Documents. Review.
Cited by
-
Whole genomes show contrasting trends of population size changes and genomic diversity for an Amazonian endemic passerine over the late quaternary.Ecol Evol. 2024 Apr 23;14(4):e11250. doi: 10.1002/ece3.11250. eCollection 2024 Apr. Ecol Evol. 2024. PMID: 38660467 Free PMC article.
-
Prioritizing Conservation Areas for the Hyacinth Macaw (Anodorhynchus hyacinthinus) in Brazil From Low-Coverage Genomic Data.Evol Appl. 2024 Nov 18;17(11):e70039. doi: 10.1111/eva.70039. eCollection 2024 Nov. Evol Appl. 2024. PMID: 39564451 Free PMC article.
-
Genomic evidence reveals high genetic diversity in a narrowly distributed species and natural hybridization risk with a widespread species in the genus Geodorum.BMC Plant Biol. 2023 Jun 14;23(1):317. doi: 10.1186/s12870-023-04285-w. BMC Plant Biol. 2023. PMID: 37316828 Free PMC article.
References
-
- Allendorf FW, Leary RF, Spruell P, Wenburg JK. 2001. The problems with hybrids: setting conservation guidelines. Trends Ecol Evol. 16:613–622.
-
- Benson JF, Patterson BR, Wheeldon TJ. 2012. Spatial genetic and morphologic structure of wolves and coyotes in relation to environmental heterogeneity in a Canis hybrid zone. Mol Ecol. 21:5934–5954. - PubMed
-
- Chambers SM, Fain SR, Fazio B, Amaral M. 2012. An account of the taxonomy of North American wolves from morphological and genetic analyses. North American Fauna 77:1–67.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

